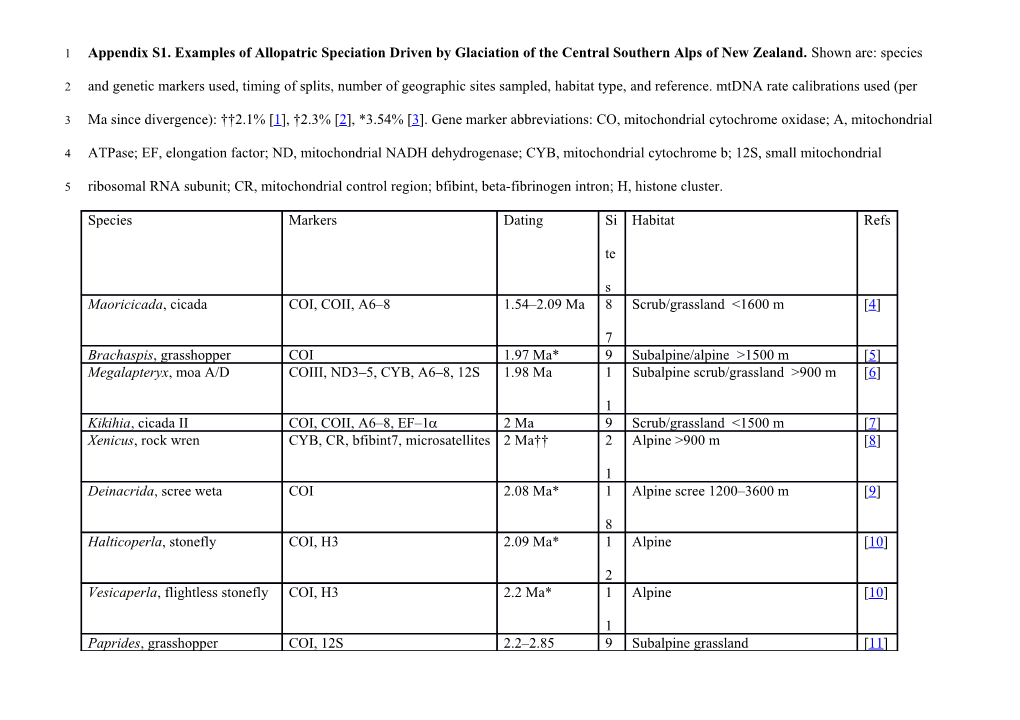
1 Appendix S1. Examples of Allopatric Speciation Driven by Glaciation of the Central Southern Alps of New Zealand. Shown are: species
2 and genetic markers used, timing of splits, number of geographic sites sampled, habitat type, and reference. mtDNA rate calibrations used (per
3 Ma since divergence): ††2.1% [1], †2.3% [2], *3.54% [3]. Gene marker abbreviations: CO, mitochondrial cytochrome oxidase; A, mitochondrial
4 ATPase; EF, elongation factor; ND, mitochondrial NADH dehydrogenase; CYB, mitochondrial cytochrome b; 12S, small mitochondrial
5 ribosomal RNA subunit; CR, mitochondrial control region; bfibint, beta-fibrinogen intron; H, histone cluster.
Species Markers Dating Si Habitat Refs
te
s Maoricicada, cicada COI, COII, A6–8 1.54–2.09 Ma 8 Scrub/grassland <1600 m [4]
7 Brachaspis, grasshopper COI 1.97 Ma* 9 Subalpine/alpine >1500 m [5] Megalapteryx, moa A/D COIII, ND3–5, CYB, A6–8, 12S 1.98 Ma 1 Subalpine scrub/grassland >900 m [6]
1 Kikihia, cicada II COI, COII, A6–8, EF–1 2 Ma 9 Scrub/grassland <1500 m [7] Xenicus, rock wren CYB, CR, bfibint7, microsatellites 2 Ma†† 2 Alpine >900 m [8]
1 Deinacrida, scree weta COI 2.08 Ma* 1 Alpine scree 1200–3600 m [9]
8 Halticoperla, stonefly COI, H3 2.09 Ma* 1 Alpine [10]
2 Vesicaperla, flightless stonefly COI, H3 2.2 Ma* 1 Alpine [10]
1 Paprides, grasshopper COI, 12S 2.2–2.85 9 Subalpine grassland [11] Ma*† Holcoperla, flightless stonefly COI, H3 2.48 Ma* 1 Alpine [10]
8 Apteryoperla, flightless stonefly COI, H3 2.51 Ma* 1 Alpine [10]
3 Cristaperla, stonefly COI, H3 2.57 Ma* 1 Alpine [10]
8 Alpinacris, grasshopper 12S 2.6 Ma† 5 Subalpine/alpine grassland [11] 6
7
2 8 References
9 1. Weir, J.T. and Schluter, D. (2008) Calibrating the avian molecular clock. Mol. Ecol. 17, 2321-2328
10 2. Brower, A.V.Z. (1994) Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from
11 patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. U.S.A. 91, 6491-6495
12 3. Papadopoulou, A., et al. (2010) Revisiting the insect mitochondrial molecular clock: the mid-Aegean Trench calibration. Mol. Biol. Evol.
13 27, 1659-1672
14 4. Hill, K.B.R., et al. (2009) Surviving glacial ages within the Biotic Gap: phylogeography of the New Zealand cicada Maoricicada
15 campbelli. J. Biogeogr. 36, 675-692
16 5. Trewick, S.A. (2001) Identity of an endangered grasshopper (Acrididae: Brachaspis): taxonomy, molecules and conservation. Conserv.
17 Genet. 2, 233-243
18 6. Bunce, M., et al. (2009) The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc. Natl. Acad.
19 Sci. U.S.A. 106, 20646-20651
3 20 7. Marshall, D.C., et al. (2008) Steady Plio-Pleistocene diversification and a 2-million-year sympatry threshold in a New Zealand cicada.
21 Mol. Phylogenet. Evol. 48, 1054-1066
22 8. Weston, K.A. and Robertson, B.C. (2015) Population structure within an alpine archipelago: strong signature of past climate change in
23 the New Zealand rock wren (Xenicus gilviventris). Mol. Ecol. 24, 4778-4794
24 9. Trewick, S.A., et al. (2000) Phylogeographical pattern correlates with Pliocene mountain building in the alpine scree weta (Orthoptera,
25 Anostostomatidae). Mol. Ecol. 9, 657-666
26 10. McCulloch, G.A., et al. (2010) Onset of glaciation drove simultaneous vicariant isolation of alpine insects in New Zealand. Evolution 64,
27 2033-2043
28 11. Trewick, S.A. and Wallis, G.P. (2001) Bridging the "beech-gap": New Zealand invertebrate phylogeography implicates Pleistocene
29 glaciation and Pliocene isolation. Evolution 55, 2170-2180
30
4