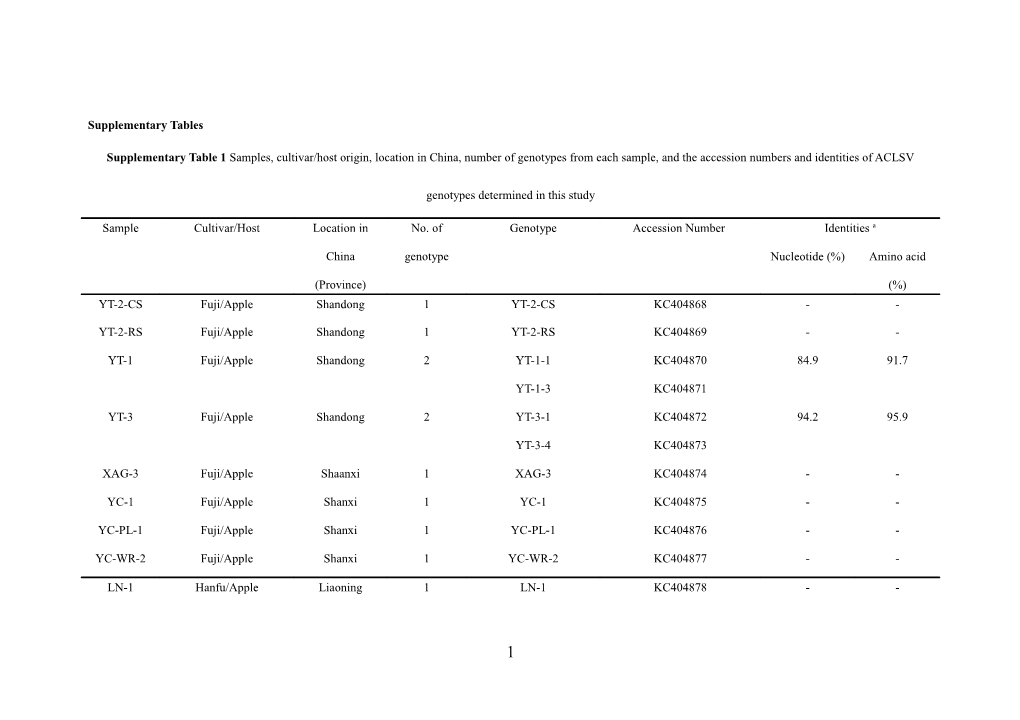
Supplementary Tables
Supplementary Table 1 Samples, cultivar/host origin, location in China, number of genotypes from each sample, and the accession numbers and identities of ACLSV
genotypes determined in this study
Sample Cultivar/Host Location in No. of Genotype Accession Number Identities a
China genotype Nucleotide (%) Amino acid
(Province) (%) YT-2-CS Fuji/Apple Shandong 1 YT-2-CS KC404868 - -
YT-2-RS Fuji/Apple Shandong 1 YT-2-RS KC404869 - -
YT-1 Fuji/Apple Shandong 2 YT-1-1 KC404870 84.9 91.7
YT-1-3 KC404871
YT-3 Fuji/Apple Shandong 2 YT-3-1 KC404872 94.2 95.9
YT-3-4 KC404873
XAG-3 Fuji/Apple Shaanxi 1 XAG-3 KC404874 - -
YC-1 Fuji/Apple Shanxi 1 YC-1 KC404875 - -
YC-PL-1 Fuji/Apple Shanxi 1 YC-PL-1 KC404876 - -
YC-WR-2 Fuji/Apple Shanxi 1 YC-WR-2 KC404877 - -
LN-1 Hanfu/Apple Liaoning 1 LN-1 KC404878 - -
1 LN-1-1 Fuji/Apple Liaoning 1 LN-1-1 KC404879 - -
LN-1-2 Fuji/Apple Liaoning 1 LN-1-2 KC404880 - -
LN-2-2 Fuji/Apple Liaoning 2 LN-2-2-1 KC404881 93.5 96.4
LN-2-2-2 KC404882
SMX-1 Fuji/Apple Henan 1 SMX-1 KC404883 - -
SMX-2 Fuji/Apple Henan 2 SMX-2-3 KC404884 85.9 94.8
SMX-2-4 KC404885
SMX-3 Fuji/Apple Henan 2 SMX-3-3 KC404886 92.1 98.5
SMX-3-4 KC404887
SMX-4 Fuji/Apple Henan 2 SMX-4-1 KC404888 88.1 94.8
SMX-4-2 KC404889
BY-3 Golden Delicious/Apple Gansu 2 BY-3-2 KC404890 99.5 99.5
BY-3-3 KC404891
BY-4 Fuji/Apple Gansu 2 BY-4-1 KC404892 88.8 96.7
BY-4-2 KC404893
BY-5 Qingguan/Apple Gansu 2 BY-5-2 KC404894 99.7 100.0
BY-5-3 KC404895
2 BY-6 Golden Delicious/Apple Gansu 2 BY-6-2 KC404896 92.7 96.7
BY-6-3 KC404897
TS-1 Fuji/Apple Gansu 1 TS-1 KC404898 - -
TS-2 Fuji/Apple Gansu 4 TS-2-1 KC404899 85.1-99.1 91.7-97.9
TS-2-2 KC404900
TS-2-3 KC404901
TS-2-4 KC404902
TS-4 Fuji/Apple Gansu 2 TS-4-1 KC404903 84.2 93.3
TS-4-2 KC404904
ZZ-2 Fuji/Apple Henan 1 ZZ-2 KC404905 - -
ZZ-3 Fuji/Apple Henan 1 ZZ-3 KC404906 - -
BJ-1 Fuji/Apple Beijing 1 BJ-1 KC404907 - -
BJ-2 Fuji/Apple Beijing 1 BJ-2 KC404908 - -
LF-2 Fuji/Apple Beijing 1 LF-2 KC404909 - -
HT-1 Unknown/Crabapple Beijing 1 HT-1 KC404910 - -
SHZ-1 Fuji/Apple Beijing 1 SHZ-1 KC404911 - -
SHZ-2 Fuji/Apple Beijing 1 SHZ-2 KC404912 - -
3 SHZ-3 Fuji/Apple Beijing 1 SHZ-3 KC404913 - -
SHZ-4 Fuji/Apple Beijing 1 SHZ-4 KC404914 - -
SHZ-5 Fuji/Apple Beijing 1 SHZ-5 KC404915 - -
SHZ-6 Fuji/Apple Beijing 1 SHZ-6 KC404916 - -
SHZ-7 Fuji/Apple Beijing 1 SHZ-7 KC404917 - -
SHZ-8 Fuji/Apple Beijing 1 SHZ-8 KC404918 - -
SHZ-10 Fuji/Apple Beijing 1 SHZ-10 KC404919 - -
MDJ-11 Long Feng /Apple Heilongjiang 1 MDJ-11 KC404920 - -
NX-1 Red delicious /Apple Ningxia 1 NX-1 KC404921 - -
NX-6 Starkrimson/Apple Ningxia 1 NX-6 KC404922 - - a Nucleotide and amino acid identities between different genotypes in the same sample.
4 Supplementary Table 2 Information on ACLSV sequences downloaded from GenBank
5 GenBank Host Geographical Genome Reference Isolate Accession no. Origin region
HQ398252.1 Peach Slovakia MP+CP Unpublished SK-BUD-pe HQ398250.1 Apple Latvia MP+CP Unpublished LV-mNP001 HQ398253.1 Plum Slovakia MP+CP Unpublished SK-92-pl HQ398251.1 Apple Latvia MP+CP Unpublished LV-mBDz007 NC_001409.1 Plum France Full-length 2 P863 JN849009.1 Peach China CP 7 S5 JN849007.1 Peach China CP 7 S3 JN849005.1 Peach China CP 7 Y3 JN849003.1 Peach China CP 7 Y1 JN849001.1 Peach China CP 7 G4 JN848999.1 Peach China CP 7 Z3 JN848997.1 Peach China CP 7 Z1 JN848995.1 Peach China CP 7 ST4 JN848993.1 Peach China CP 7 ST2 JN848991.1 Peach China CP 7 SG8 JN848989.1 Peach China CP 7 SQ5 JN848987.1 Peach China CP 7 SQ2 JN848985.1 Peach China CP 7 HL1 JN848983.1 Peach China CP 7 HB4 JN848979.1 Peach China CP 7 HS6 JN848977.1 Peach China CP 7 HS4 JN848975.1 Peach China CP 7 HS2 JN848973.1 Peach China CP 7 HC1 JN848971.1 Peach China CP 7 BL1 JN849010.1 Peach China CP 7 S6 JN849008.1 Peach China CP 7 S4 JN849006.1 Peach China CP 7 S1 JN849004.1 Peach China CP 7 Y2 JN849002.1 Peach China CP 7 G6 JN849000.1 Peach China CP 7 G2 JN848998.1 Peach China CP 7 Z2 JN848996.1 Peach China CP 7 F1 JN848994.1 Peach China CP 7 ST3 JN848992.1 Peach China CP 7 ST1 JN848990.1 Peach China CP 7 SQ6 JN848988.1 Peach China CP 7 SQ3 JN848986.1 Peach China CP 7 SQ1 JN848984.1 Peach China CP 7 HB5 JN848982.1 Peach China CP 7 HB3 JN848980.1 Peach China CP 7 HB1 JN848978.1 Peach China CP 7 HS5 JN848976.1 Peach China CP 7 HS3 JN848974.1 Peach China CP 7 HS1 JN848981.1 Peach China CP 7 HB2 JN848972.1 Peach China CP 7 BN1 HQ670737.1 Apple Latvia MP+CP Unpublished LV-mBDz007-2 FR873735.1 Apple India CP Unpublished --- GU328004.1 Pear China CP 16 PP64 GU328002.1 Pear China CP 16 PP61 GU328000.1 Pear China CP 16 PP57 GU327998.1 Pear China CP 16 PP55 GU327996.1 Pear China CP 16 PP43 GU327994.1 Pear China CP 16 PP36 GU327992.1 Pear China CP 16 PP24 GU327990.1 Pear China CP 16 PP20 GU327988.1 Pear China CP 16 PP15-2 GU327986.1 Pear China CP 16 PP12 GU327984.1 Pear China CP 16 PP5 GU327982.1 Plum China CP 16 PL-1 GU328003.1 Pear China CP 16 PP63 GU328001.1 Pear China CP 16 PP58 GU327999.1 Pear China CP 16 PP56 GU327997.1 Pear China CP 16 PP54 GU327995.1 Pear China CP 16 PP39 GU327993.1 Pear China CP 16 PP27 GU327991.1 Pear China CP 16 PP23 GU327989.1 Pear China CP 16 PP15-4 GU327987.1 Pear China CP6 16 PP13 GU327985.1 Pear China CP 16 PP6 GU327983.1 Plum China CP 16 PL2 GU327981.1 Peach China CP 16 PE AM709776.2 Apple India CP Unpublished BE HM352769.1 Pear Taiwan CP 17 LTS2 Note: MP, movement protein; CP, coat protein; “---” indicates relevant information not available.
Reference
1. Cai Y, Xiang BC, Xi DH, Liu SX, Du YJ (2005) Cloning and prokaryotic expression of CP gene of Apple chlorotic leaf spot virus Kuerle isolate and preparation of its specific antiserum. Journal of Agricultural Biotechnology 13:533-537 (in Chinese). 2. German S, Candresse T, Lanneau M, Huet JC, Pernollet JC, Dunez J (1990) Nucleotide sequence and genomic organization of Apple chlorotic leaf spot closterovirus. Virology 179:104-112. 3. German-Retana S, Bergey B, Delbos RP, Candresse T, Dunez J (1997) Complete nucleotide sequence of the genome of a severe cherry isolate of apple chlorotic leaf spot trichovirus (ACLSV). Arch Virol 142:833-841. 4. Jelkmann W (1996) The nucleotide sequence of a strain of Apple chlorotic leaf spot virus (ACLSV) responsible for plum pseudopox and its relation to an apple and plum bark split strain. Phytopathology 86:101. 5. Malinowski T, Komorowska B, Cieslinska M, Candresse T, Zawadzka B (1998) Characterization of SX/2, an Apple chlorotic leaf spot virus isolate showing unusual coat protein properties. Acta Hortic, 472: 43-50. 6. Marini DB, Gibson PG, Scott SW (2008) The complete nucleotide sequence of an isolate of apple chlorotic leaf spot virus from peach (Prunus persica (L.) Batch). Arch Virol 153:1003-1005. 7. Niu FQ, Pan S, Wu ZJ, Jiang DM, Li SF (2012) Complete nucleotide sequences of the genomes of two isolates of apple chlorotic leaf spot virus from peach (Prunus persica) in China. Arch Virol 157:783-786. 8. Rana T, Chandel V, Hallan V, Handa A, Thakur PD, Zaidi AA (2008) Molecular characterization of almond isolate of Apple chlorotic leaf spot virus from India. Australas Plant Pathol 3:65-67. 9. Rana T, Chandel V, Hallan V, Singh AK, Handa A, Thakur PD, Zaidi AA (2007) Molecular characterization of Apple chlorotic leaf spot virus from India. Indian J Virol 18:70-74. 10. Rana T, Chandel V, Hallan V, Zaidi AA (2007) Molecular characterization of Chuli isolate of Apple chlorotic leaf spot virus from India. J Plant Pathol 89: S72. 11. Rana T, Chandel V, Hallan V, Zaidi AA (2007) Molecular characterization of Cydonia isolate of Apple chlorotic leaf spot virus from India. Plant Pathol 57:393-393. 12. Rana T, Chandel V, Hallan V, Zaidi AA (2008) Himalayan wild cherry (Prunus cerasoides D. Don): a new host of Apple chlorotic leaf spot virus. For Pathol 38:73-77. 13. Rana T, Chandel V, Hallan V, Zaidi AA (2009) Molecular evidence for the presence of Apple chlorotic leaf spot virus in infected peach trees in India. Sci Hortic 120:296-299. 14. Rana T, Chandel V, Kumar Y, Ram R, Hallan V, Zaidi AA (2010) Molecular variability analyses of Apple chlorotic leaf spot virus capsid protein. J Bioscience 35:605-615. 15. Sato K, Yoshikawa N, Takahashi T (1993) Complete nucleotide sequence of the genome of an apple isolate of Apple chlorotic leaf spot virus. J Gen Virol 74:1927-1931. 16. Song YS, Hong N, Wang LP, Hu HJ, Tian R, Xu WX, Ding F, Wang GP (2011) Molecular and serological diversity in Apple chlorotic leaf spot virus from sand pear (Pyrus pyrifolia) in China. Eur J Plant Pathol 130:183-196. 17. Wu ZB, Ku HM, ChenYK, Chang CJ, Jan FJ (2010) Biological and molecular characterization of Apple chlorotic leaf spot virus causing chlorotic leaf spot on pear (Pyrus pyrifolia) in Taiwan. HortScience 45:1073-1078.
7 18. Yaegashi H, Isogai M, Tajima H, Sano T, Yoshikawa N (2007) Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of apple chlorotic leaf spot virus are crucial for infectivity. J Gen Virol 88:2611-2618. 19. Zheng YY, Wang GP, Hong N, Song YS, You H (2007) Partial molecular characterization of Apple chlorotic leaf spot virus from peach and apple trees and prokaryotic expression for cp gene. Acta Phytopathologica Sinica 37:356-361 (in Chinese).
8 Supplementary Table 3 Genetic distances within and among clusters based on nucleotide alignments shown in Fig. 1b for ACLSV isolates and genotypes worldwide Cluster I II III IVa (Ta Tao 5 type) (SHZ type) (P205 type) (B6 type ) I (Ta Tao 5 type) 0.114±0.009 0.392±0.032 0.394±0.031 0.379±0.030 II (SHZ type) - 0.060±0.006 0.154±0.014 0.167±0.015 III (P205 type) - - 0.090±0.008 0.159±0.014 IV (B6 type)a - - - 0.102±0.009 a, Cluster IV contained all the isolates and genotypes that were not in Clusters I, II, and III in Fig. 1b
9 Supplementary Figures and Figure legends
Supplementary Figure 1 Amino acid sequence alignment of ACLSV CP of 55 isolates from apple in China. Co-variation of amino acid at positions 40, 75, and 79 in three groups is indicated by triangles. The unique aa in positions 79, 82, and 98 for group II are indicated by a gray
10 Supplementary Figure 2 Amino acid sequence alignment of ACLSV CP of 125 isolates from China. Co-variation of aa at positions 40, 75, and 79 in four clusters is indicated by triangles
11 Supplementary Figure 3 Phylogenetic tree of non-recombinant ACLSV CP amino acid sequences of 123 isolates from China. The tree was constructed by the neighbor-joining method using MEGA5.03, with 100 as bootstrap replicates. Only bootstrap values ≥ 70% are shown. Four clusters (types Ta Tao 5, SHZ, P205 and B6) in the tree are circled. Bar, 0.02 amino acid replacements per site
12 Supplementary Figure 4 Phylogenetic tree of non-recombinant ACLSV CP amino acid sequences of 173 isolates worldwide. The tree was constructed by the neighbor-joining method using MEGA5.03, with 100 bootstrap replicates. Only bootstrap values ≥ 70% are shown. Three clusters (types Ta Tao 5, SHZ, and P205) in the tree are circled. Bar, 0.01 amino acid replacements per site
13