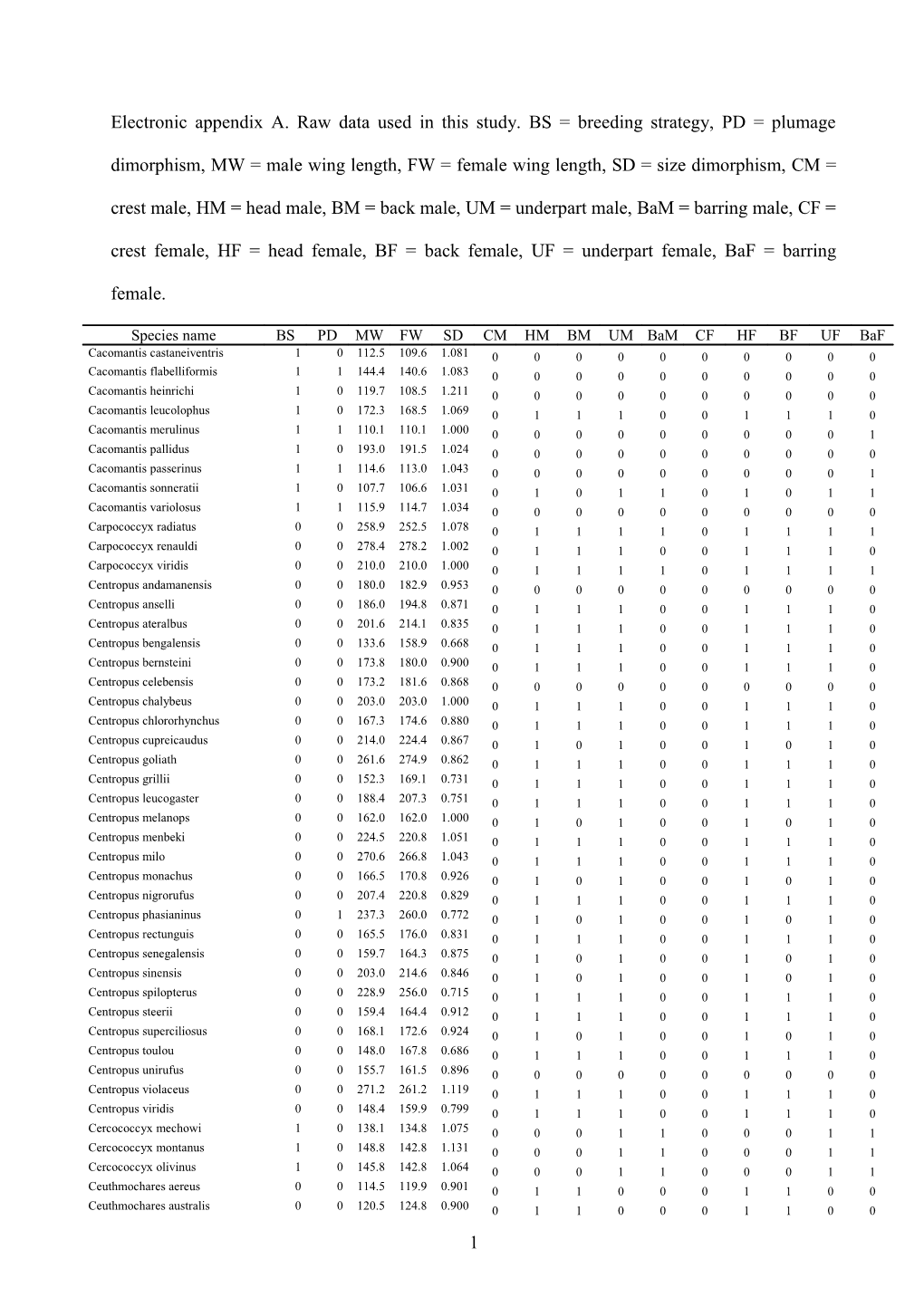
Electronic appendix A. Raw data used in this study. BS = breeding strategy, PD = plumage
dimorphism, MW = male wing length, FW = female wing length, SD = size dimorphism, CM =
crest male, HM = head male, BM = back male, UM = underpart male, BaM = barring male, CF =
crest female, HF = head female, BF = back female, UF = underpart female, BaF = barring
female.
Species name BS PD MW FW SD CM HM BM UM BaM CF HF BF UF BaF Cacomantis castaneiventris 1 0 112.5 109.6 1.081 0 0 0 0 0 0 0 0 0 0 Cacomantis flabelliformis 1 1 144.4 140.6 1.083 0 0 0 0 0 0 0 0 0 0 Cacomantis heinrichi 1 0 119.7 108.5 1.211 0 0 0 0 0 0 0 0 0 0 Cacomantis leucolophus 1 0 172.3 168.5 1.069 0 1 1 1 0 0 1 1 1 0 Cacomantis merulinus 1 1 110.1 110.1 1.000 0 0 0 0 0 0 0 0 0 1 Cacomantis pallidus 1 0 193.0 191.5 1.024 0 0 0 0 0 0 0 0 0 0 Cacomantis passerinus 1 1 114.6 113.0 1.043 0 0 0 0 0 0 0 0 0 1 Cacomantis sonneratii 1 0 107.7 106.6 1.031 0 1 0 1 1 0 1 0 1 1 Cacomantis variolosus 1 1 115.9 114.7 1.034 0 0 0 0 0 0 0 0 0 0 Carpococcyx radiatus 0 0 258.9 252.5 1.078 0 1 1 1 1 0 1 1 1 1 Carpococcyx renauldi 0 0 278.4 278.2 1.002 0 1 1 1 0 0 1 1 1 0 Carpococcyx viridis 0 0 210.0 210.0 1.000 0 1 1 1 1 0 1 1 1 1 Centropus andamanensis 0 0 180.0 182.9 0.953 0 0 0 0 0 0 0 0 0 0 Centropus anselli 0 0 186.0 194.8 0.871 0 1 1 1 0 0 1 1 1 0 Centropus ateralbus 0 0 201.6 214.1 0.835 0 1 1 1 0 0 1 1 1 0 Centropus bengalensis 0 0 133.6 158.9 0.668 0 1 1 1 0 0 1 1 1 0 Centropus bernsteini 0 0 173.8 180.0 0.900 0 1 1 1 0 0 1 1 1 0 Centropus celebensis 0 0 173.2 181.6 0.868 0 0 0 0 0 0 0 0 0 0 Centropus chalybeus 0 0 203.0 203.0 1.000 0 1 1 1 0 0 1 1 1 0 Centropus chlororhynchus 0 0 167.3 174.6 0.880 0 1 1 1 0 0 1 1 1 0 Centropus cupreicaudus 0 0 214.0 224.4 0.867 0 1 0 1 0 0 1 0 1 0 Centropus goliath 0 0 261.6 274.9 0.862 0 1 1 1 0 0 1 1 1 0 Centropus grillii 0 0 152.3 169.1 0.731 0 1 1 1 0 0 1 1 1 0 Centropus leucogaster 0 0 188.4 207.3 0.751 0 1 1 1 0 0 1 1 1 0 Centropus melanops 0 0 162.0 162.0 1.000 0 1 0 1 0 0 1 0 1 0 Centropus menbeki 0 0 224.5 220.8 1.051 0 1 1 1 0 0 1 1 1 0 Centropus milo 0 0 270.6 266.8 1.043 0 1 1 1 0 0 1 1 1 0 Centropus monachus 0 0 166.5 170.8 0.926 0 1 0 1 0 0 1 0 1 0 Centropus nigrorufus 0 0 207.4 220.8 0.829 0 1 1 1 0 0 1 1 1 0 Centropus phasianinus 0 1 237.3 260.0 0.772 0 1 0 1 0 0 1 0 1 0 Centropus rectunguis 0 0 165.5 176.0 0.831 0 1 1 1 0 0 1 1 1 0 Centropus senegalensis 0 0 159.7 164.3 0.875 0 1 0 1 0 0 1 0 1 0 Centropus sinensis 0 0 203.0 214.6 0.846 0 1 0 1 0 0 1 0 1 0 Centropus spilopterus 0 0 228.9 256.0 0.715 0 1 1 1 0 0 1 1 1 0 Centropus steerii 0 0 159.4 164.4 0.912 0 1 1 1 0 0 1 1 1 0 Centropus superciliosus 0 0 168.1 172.6 0.924 0 1 0 1 0 0 1 0 1 0 Centropus toulou 0 0 148.0 167.8 0.686 0 1 1 1 0 0 1 1 1 0 Centropus unirufus 0 0 155.7 161.5 0.896 0 0 0 0 0 0 0 0 0 0 Centropus violaceus 0 0 271.2 261.2 1.119 0 1 1 1 0 0 1 1 1 0 Centropus viridis 0 0 148.4 159.9 0.799 0 1 1 1 0 0 1 1 1 0 Cercococcyx mechowi 1 0 138.1 134.8 1.075 0 0 0 1 1 0 0 0 1 1 Cercococcyx montanus 1 0 148.8 142.8 1.131 0 0 0 1 1 0 0 0 1 1 Cercococcyx olivinus 1 0 145.8 142.8 1.064 0 0 0 1 1 0 0 0 1 1 Ceuthmochares aereus 0 0 114.5 119.9 0.901 0 1 1 0 0 0 1 1 0 0 Ceuthmochares australis 0 0 120.5 124.8 0.900 0 1 1 0 0 0 1 1 0 0 1 Chrysococcyx basalis 1 0 102.7 100.7 1.061 0 1 0 1 1 0 1 0 1 1 Chrysococcyx caprius 1 4 111.4 112.0 0.984 0 1 1 1 0 0 0 0 0 1 Chrysococcyx cupreus 1 3 105.9 100.6 1.167 0 1 1 1 0 0 0 1 1 1 Chrysococcyx flavigularis 1 3 94.6 94.8 0.994 0 1 0 1 1 0 0 0 0 1 Chrysococcyx klaas 1 3 96.0 94.7 1.042 0 1 1 1 1 0 0 1 1 1 Chrysococcyx lucidus 1 1 106.3 105.6 1.021 0 1 1 1 1 0 0 1 1 1 Chrysococcyx maculatus 1 3 108.8 109.1 0.992 0 1 1 1 1 0 0 1 1 1 Chrysococcyx megarhynchus 1 2 97.8 99.1 0.961 0 0 0 0 0 0 0 0 0 1 Chrysococcyx meyeri 1 2 88.6 89.1 0.983 0 1 1 1 1 0 1 1 1 1 Chrysococcyx minutillus 1 1 94.1 92.9 1.036 0 1 1 1 1 0 1 0 1 1 Chrysococcyx osculans 1 0 118.4 116.9 1.039 0 1 0 1 0 0 1 0 1 0 Chrysococcyx ruficollis 1 1 94.8 95.5 0.978 0 0 0 1 1 0 0 0 1 1 Chrysococcyx xanthorhynchus 1 4 100.2 99.6 1.018 0 1 1 1 1 0 0 0 1 1 Clamator coromandus 1 0 161.3 160.1 1.023 1 1 1 1 0 1 1 1 1 0 Clamator glandarius 1 0 199.0 194.1 1.115 1 1 0 1 0 1 1 0 1 0 Clamator jacopica 1 0 147.2 148.5 0.974 1 1 1 1 0 1 1 1 1 0 Clamator jacoserratus 1 0 153.9 152.4 1.030 1 1 1 1 0 1 1 1 1 0 Clamator levaillantii 1 0 173.9 174.2 0.995 1 1 1 1 0 1 1 1 1 0 Coccycua cinerea 0 0 106.8 109.6 0.925 0 1 0 1 0 0 1 0 1 0 Coccycua minuta 0 0 105.2 105.0 1.006 0 1 0 0 0 0 1 0 0 0 Coccycua pumila 0 0 102.2 102.3 0.997 0 1 0 1 0 0 1 0 1 0 Coccyzus americanus 0 0 138.8 144.2 0.892 0 1 0 1 0 0 1 0 1 0 Coccyzus erythropthalmus 0 0 135.2 139.2 0.916 0 1 0 1 0 0 1 0 1 0 Coccyzus euleri 0 0 128.2 133.0 0.896 0 1 0 1 0 0 1 0 1 0 Coccyzus ferrugineus 0 0 131.5 134.2 0.941 0 0 0 0 0 0 0 0 0 0 Coccyzus lansbergi 0 0 113.9 113.2 1.019 0 0 0 0 0 0 0 0 0 0 Coccyzus longirostris 0 0 134.3 137.8 0.926 0 1 0 1 0 0 1 0 1 0 Coccyzus melacoryphus 0 0 117.9 119.2 0.968 0 0 0 0 0 0 0 0 0 0 Coccyzus merlini 0 0 174.0 180.0 0.903 0 1 0 1 0 0 1 0 1 0 Coccyzus minor 0 0 136.5 136.9 0.991 0 0 0 0 0 0 0 0 0 0 Coccyzus pluvialis 0 0 177.4 190.5 0.808 0 1 0 1 0 0 1 0 1 0 Coccyzus rufigularis 0 0 171.6 180.8 0.855 0 1 0 0 0 0 1 0 0 0 Coccyzus vetula 0 0 123.8 128.8 0.888 0 1 0 1 0 0 1 0 1 0 Coccyzus vieilloti 0 0 128.5 130.4 0.957 0 1 0 1 0 0 1 0 1 0 Coua caerulea 0 0 195.2 199.5 0.937 1 1 1 1 0 1 1 1 1 0 Coua coquereli 0 0 144.0 147.8 0.925 0 1 0 0 0 0 1 0 0 0 Coua cristata 0 0 139.2 145.9 0.868 1 1 1 1 0 1 1 1 1 0 Coua cursor 0 0 129.8 134.7 0.895 0 1 0 0 0 0 1 0 0 0 Coua delalandei 0 0 0 1 1 1 0 0 1 1 1 0 Coua gigas 0 0 220.7 213.5 1.105 0 1 0 0 0 0 1 0 0 0 Coua reynaudii 0 0 136.9 139.1 0.953 0 1 1 1 0 0 1 1 1 0 Coua ruficeps 0 0 167.5 167.9 0.993 0 1 1 0 0 0 1 1 0 0 Coua serriana 0 0 164.5 168.0 0.939 0 1 0 0 0 0 1 0 0 0 Coua verreauxi 0 0 133.2 130.3 1.068 1 1 1 1 0 1 1 1 1 0 Crotophaga ani 0 0 141.8 145.9 0.918 0 1 1 1 0 0 1 1 1 0 Crotophaga major 0 0 202.6 201.0 1.024 0 1 1 1 0 0 1 1 1 0 Crotophaga sulcirostris 0 0 134.7 131.2 1.082 0 1 1 1 0 0 1 1 1 0 Cuculus canorus 1 2 221.0 210.0 1.166 0 0 0 1 1 0 0 0 1 1 Cuculus clamosus 1 1 167.9 160.8 1.138 0 1 1 1 1 0 1 1 1 1 Cuculus crassirostris 1 0 206.1 198.0 1.128 0 0 0 1 1 0 0 0 1 1 Cuculus gularis 1 3 215.3 205.3 1.153 0 0 0 1 1 0 0 0 1 1 Cuculus micropterus 1 2 195.3 193.6 1.027 0 0 0 1 1 0 0 0 1 1 Cuculus poliocephalus 1 1 149.8 145.8 1.085 0 0 0 1 1 0 0 0 1 1 Cuculus rochii 1 0 166.1 159.7 1.125 0 0 0 1 1 0 0 0 1 1 Cuculus saturatus 1 2 183.4 175.4 1.143 0 0 0 1 1 0 0 0 1 1 Cuculus solitarius 1 1 172.0 169.0 1.054 0 0 0 1 1 0 0 0 1 1 Dasylophus cumingi 0 0 156.9 159.0 0.961 0 1 1 0 0 0 1 1 0 0 2 Dasylophus superciliosus 0 0 150.8 156.5 0.895 1 1 1 1 0 1 1 1 1 0 Dromococcyx pavonicus 1 0 134.4 133.8 1.014 1 0 0 1 0 1 0 0 1 0 Dromococcyx phasianellus 1 0 166.2 162.8 1.064 1 0 0 1 0 1 0 0 1 0 Eud scocyano 1 4 217.2 214.6 1.037 0 1 1 1 0 0 0 0 0 1 Eudynamys scolopacea 1 4 194.8 188.6 1.102 0 1 1 1 0 0 0 0 1 1 Geococcyx californianus 0 1 178.3 170.7 1.140 1 0 0 1 1 1 0 0 1 1 Geococcyx velox 0 0 145.9 142.1 1.082 1 0 0 1 0 1 0 0 1 0 Guira guira 0 0 178.5 174.3 1.074 1 1 0 1 0 1 1 0 1 0 Hierococcyx bocki 1 0 186.3 189.7 0.947 0 0 0 1 1 0 0 0 1 1 Hierococcyx fugax 1 0 174.3 173.4 1.016 0 0 0 1 1 0 0 0 1 1 Hierococcyx hyperythrus 1 0 199.0 196.0 1.047 0 0 0 0 0 0 0 0 0 0 Hierococcyx nisicolor 1 0 178.3 181.0 0.956 0 0 0 1 1 0 0 0 1 1 Hierococcyx pectoralis 1 0 174.8 174.2 1.010 0 0 0 0 0 0 0 0 0 0 Hierococcyx sparverioides 1 0 236.0 226.7 1.128 0 0 0 1 1 0 0 0 1 1 Hierococcyx vagans 1 0 145.0 143.1 1.040 0 1 0 1 1 0 1 0 1 1 Hierococcyx varius 1 0 200.5 191.4 1.150 0 0 0 1 1 0 0 0 1 1 Microdynamis parva 1 3 105.4 101.5 1.120 0 1 0 0 0 0 0 0 0 0 Morococcyx erythropygus 0 0 98.7 96.9 1.057 0 1 0 1 0 0 1 0 1 0 Neomorphus geoffroyi 0 0 172.4 170.1 1.041 1 1 1 1 0 1 1 1 1 0 Neomorphus pucheranii 0 0 171.7 165.8 1.111 1 1 1 1 0 1 1 1 1 0 Neomorphus radiolosus 0 0 166.0 167.0 0.982 1 1 0 1 1 1 1 0 1 1 Neomorphus rufipennis 0 1 169.5 164.3 1.098 1 1 1 1 0 1 1 1 1 0 Pachycoccyx audeberti 1 0 223.3 220.8 1.034 0 0 0 1 0 0 0 0 1 0 Phaenicophaeus curvirostris 0 0 169.1 171.5 0.959 0 1 1 0 0 0 1 1 0 0 Phaenicophaeus diardi 0 0 131.9 130.9 1.023 0 1 1 0 0 0 1 1 0 0 Phaenicophaeus pyrrhocephalus 0 0 154.7 157.0 0.957 0 1 1 1 0 0 1 1 1 0 Phaenicophaeus sumatranus 0 0 139.4 138.6 1.017 0 1 1 0 0 0 1 1 0 0 Phaenicophaeus tristis 0 0 163.3 161.2 1.040 0 1 1 0 0 0 1 1 0 0 Phaenicophaeus viridirostris 0 0 135.0 131.0 1.094 0 1 1 0 0 0 1 1 0 0 Piaya cayana 0 0 146.1 145.7 1.008 0 1 0 1 0 0 1 0 1 0 Piaya melanogaster 0 0 135.8 135.4 1.009 0 1 1 1 0 0 1 1 1 0 Rhamphococcyx calyorhynchus 0 0 175.4 177.8 0.960 0 1 1 0 0 0 1 1 0 0 Rhinorta chlorophaeus 0 3 114.6 115.9 0.967 0 1 0 0 0 0 1 1 0 0 Scythrops novaehollandiae 1 0 353.9 346.8 1.063 0 1 0 0 1 0 1 0 0 1 Surniculus dicruroides 1 0 137.2 135.9 1.031 0 1 1 1 0 0 1 1 1 0 Surniculus lugubris 1 0 121.6 120.9 1.017 0 1 1 1 0 0 1 1 1 0 Surniculus musschenbroeki 1 0 131.1 128.6 1.059 0 1 1 1 0 0 1 1 1 0 Surniculus velutinus 1 0 117.2 117.1 1.003 0 1 1 1 0 0 1 1 1 0 Taccocua leschenaultii 0 0 157.6 153.5 1.082 0 1 0 0 0 0 1 0 0 0 Tapera naevia 1 0 118.8 113.4 1.156 1 0 0 1 0 1 0 0 1 0 Urodynamys taitensis 1 0 189.0 185.5 1.058 0 0 0 1 0 0 0 0 1 0 Zanclostomus javanicus 0 0 144.3 147.3 0.940 0 1 1 1 0 0 1 1 1 0
3 Electronic appendix B. 171 taxon parsimony tree from Sorenson & Payne (2005) with maximum likelihood estimates of branch lengths under a GTR+I+G model of sequence evolution. Branch lengths based on 1959 aligned nucleotide positions, comprising all of the mitochondrial ND2 gene plus well-aligned portions of the small subunit (12S) rRNA gene and portions of the tRNA genes flanking both ND2 and 12S. Due to ambiguous alignment and different numbers of nucleotides in different taxa, “gap regions” comprising 263 alignment positions were excluded for the estimation of branch lengths.
Model parameters:
Tree 1 ------ln L 57653.78014 Base frequencies: A 0.391217 C 0.355078 G 0.062657 T 0.191049 Rate matrix R: AC 0.33264 AG 8.64735 AT 0.41544 CG 0.25186 CT 5.92021 GT 1.00000 P_inv 0.355925 Shape 0.629067
Translate 1 JGG1162_Cu_saturatus_saturatus, 2 CM3840_Cu_saturatus_optatus, 3 T1107_Cu_saturatus_optatus, 4 A830_Cu_canorus, 5 A1201_Cu_canorus_subtelephonus, 6 BKS2197_Cu_canorus, 7 SMG8019_Cu_rochii, 8 US527286_Cu_gularis, 9 WLK132_Cu_micropterus, 10 AMNH298824_Cu_crassirostris, 11 T1295_Cu_poliocephalus, 12 PRS2426_Cu_poliocephalus, 13 GAV463_Cu_solitarius, 14 GAV640_Cu_c_clamosus, 15 WLK410_Hi_nisicolor, 4 16 WLK500_Hi_nisicolor, 17 WLK484_Hi_fugax, 18 CM2047_Hi_pectoralis, 19 A947_Hi_hyperythrus, 20 JGG1102_Hi_sparverioides, 21 A1569_Hi_varius, 22 AMNH648292_Hi_bocki, 23 US534592_Hi_vagans, 24 WFVZ37594_Su_lugubris, 25 US571628_Su_musschenbroeki, 26 F286173_Su_velutinus, 27 F221725_Su_dicruroides, 28 ypm50330_Cercococcyx_olivinus, 29 TPG966_Cercococcyx_montanus, 30 RWD23768_Cercococcyx_mechowii, 31 ATP92.048_Ca_v_sepulcralis, 32 QM26747_Ca_v_variolosus, 33 ypm74832_Ca_v_heinrichi, 34 A483_Ca_merulinus, 35 A482_Ca_sonneratii, 36 QM27099_Ca_castaneiventris, 37 QM30248_Ca_flabelliformis, 38 CES36_Ca_pallidus, 39 mvz149109_Ca_leucolophus, 40 QM28434_Ch_minutillus_poecilurus, 41 QM29712_Ch_minutillus_barnardi, 42 A1575_Ch_minutillus_crassirostris, 43 ypm89143_Ch_meyeri, 44 221890_Ch_lucidus_layardi, 45 CES065_Ch_lucidus_plagosus, 46 ypm74870_Ch_ruficollis, 47 ANWL28619_Ch_osculans, 48 SAR7138_Ch_basalis, 49 ypm75000_Ch_megarhynchus, 50 A850_Ch_cupreus, 51 B39432_Ch_cupreus, 52 CGS5242_Ch_flavigularis, 53 ALP76_Ch_klaas, 54 SAR6914_Ch_caprius, 55 JCK1654_Ch_caprius, 56 ypm39592_Ch_maculatus, 57 WLK424_Ch_xanthorhynchus, 58 A1220_Urodynamis_taitensis, 59 QM29923_Scythrops_novaehollandiae, 60 CES160_Eudynamys_s_alberti, 61 mcz39733_Eudynamys_s_melanorhynchus, 62 A1171_Eudynamys_s_scolopacea, 63 114159_Microdynamis_parva_grisescens, 64 PB1089.89_Pachycoccyx_audeberti, 65 ANS5563_Co_merlini, 66 A577_Co_longirostris,
5 67 B11323_Co_vieilloti, 68 AMNH155237_Co_vetula, 69 UF26233_Co_pluvialis, 70 AMNH164128_Co_rufigularis, 71 ANS5233_Co_lansbergi, 72 T1012_Co_erythropthalmus, 73 T453_Co_americanus, 74 ANS4661_Co_euleri, 75 T627_Co_minor, 76 CM123814_Co_ferrugineus, 77 ANS5958_Co_melacoryphus, 78 B14529_Piaya_cayana, 79 B10769_Piaya_melanogaster, 80 CU35069_Coccycua_pumilus, 81 LSU6833_Coccycua_cinereus, 82 B1179_Coccycua_minuta, 83 A1660_Clamator_jacobinus_pica, 84 A641_Clamator_levaillantii, 85 A1034_Clamator_glandarius, 86 A481_Clamator_coromandus, 87 228024_Dasylophus_superciliosus, 88 CM636_Dasylophus_cumingi, 89 A1584_Rhamphococcyx_calyorhynchus, 90 ANSP139384_Ph_d_diardi, 91 OU15102_Ph_t_longicaudatus, 92 F229803_Ph_viridirostris, 93 F278258_Ph_pyrrhocephalus, 94 ypm51093_Ph_sumatranus, 95 T1576_Ph_curvirostris, 96 A486_Zanclostomus_javanicus, 97 BD4744_Zanclostomus_javanicus, 98 MSU4063_Taccocua_leschenaultii, 99 PB14_Ceuthmochares_a_aereus, 100 A1068_Ceuthmochares_a_australis, 101 OU15103_Rhinortha_c_chlorophaea, 102 QM29481_Ce_p_phasianus, 103 ypm89146_Ce_bernsteini, 104 Cop101_Ce_violaceus, 105 CM4184_Ce_bengalensis_javanensis, 106 CM4187_Ce_viridis, 107 A1051_Ce_grillii, 108 US571638_Ce_goliath, 109 SMG8016_Ce_toulou, 110 A1172_Ce_sinensis, 111 A1975_Ce_andamanensis, 112 US219367_Ce_nigrorufus, 113 A764_Ce_cupreicaudus, 114 VGR65_Ce_superciliosus_burchellii, 115 ypm50358_Ce_monachus, 116 A377_Ce_senegalensis, 117 A1572_Ce_l_leucogaster,
6 118 ypm50356_Ce_anselli, 119 mcz270146_Ce_c_celebensis, 120 ypm34670_Ce_steerii, 121 AMNH628149_Ce_rectunguis, 122 CM3099_Ce_melanops, 123 mcz184954_Ce_chlororhynchus, 124 DM68328_Ce_unirufus, 125 ypm74879_Ce_menbeki, 126 ANSP132995_Ce_chalybeus, 127 Cop102_Ce_ateralbus, 128 CEF263_Ce_milo, 129 TSS4770_Coua_serriana, 130 mcz84298_Coua_delalandei, 131 SMG7813_Coua_gigas, 132 ypm69417_Coua_coquereli, 133 SMG4092_Coua_cursor, 134 SMG4217_Coua_reynaudii, 135 JCR51_Coua_ruficeps_olivaceiceps, 136 SMG4279_Coua_caerulea, 137 AMNH628959_Coua_verreauxi, 138 SMG7814_Coua_cristata, 139 T1578_Carpococcyx_renauldi, 140 A1586_Carpococcyx_radiatus, 141 A1578_Carpococcyx_viridis, 142 AMNH278613_Neomorphus_g_squamiger, 143 B2319_Neomorphus_g_geoffroyi, 144 ypm54524_Neomorphus_radiolosus, 145 ANS7727_Neomorphus_rufipennis, 146 LSU4983_Neomorphus_pucheranii, 147 A353_Geococcyx_californianus, 148 DAB1518_Geococcyx_velox, 149 DAB1569_Morococcyx_erythropygus, 150 B11245_Dromococcyx_phasianellus, 151 B11072_Dromococcyx_pavoninus, 152 B15026_Tapera_naevia, 153 B11350_Crotophaga_ani, 154 DAB988_Crotophaga_sulcirostris, 155 ANS3228_Crotophaga_major, 156 B6625_Guira_guira, 157 T1577_Guira_guira, 158 SMSH_Smithornis_sharpei, 159 SAPH_Sayornis_phoebe, 160 VICH_Vidua_chalybeata, 161 A525_Lanius_senator, 162 T727_Urocolius_macrourus, 163 T988_Colius_striatus, 164 B10754_Opisthocomus_hoazin, 165 B9960_Opisthocomus_hoazin, 166 A573_Musophaga_violacea, 167 A794_Crinifer_piscator, 168 92.118_Neophema_elegans,
7 169 SMG2520_Nandayus_nenday, 170 T1067_Treron_sieboldii, 171 T272_Columba_leucocephala ; tree PAUP_1 = [&U] (((((((((((((((((((((1:0.001145,5:0.001134):0.001182,2:0.004011):0.000519,4:0.001120):0.001153 ,(3:0.002270,6:0.000551):0.003996):0.003765, (7:0.003157,8:0.009474):0.002276):0.007988,9:0.015090):0.005876,10:0.021275):0.003652, (11:0.001182,12:0.000480):0.023217):0.006818,13:0.024953):0.022653,14:0.048951):0.012034, ((((((15:0.001116,16:0.000554):0.002741,17:0.006431):0.001816,18:0.006901):0.007615,19:0.0 15111):0.012628, ((20:0.005368,21:0.008332):0.005098,22:0.008471):0.026292):0.007047,23:0.035314):0.016718 ):0.003818, ((((24:0.008639,25:0.006347):0.002140,26:0.011209):0.010603,27:0.012701):0.039695, ((28:0.014483,29:0.011741):0.024061,30:0.034752):0.018522):0.000489):0.021107, ((((((31:0.008563,32:0.011000):0.002198,33:0.008270):0.019946,34:0.043122):0.019841,35:0.0 59532):0.004623,(36:0.014039,37:0.020467):0.031965):0.007090, (38:0.060925,39:0.021997):0):0.024720):0.034404, ((((((((40:0,41:0):0.003634,42:0.004557):0.014638,43:0.012522):0.003605, ((44:0.005596,45:0.002269):0.014272,46:0.008751):0.006125):0,47:0.033209):0.010447,48:0.0 64941):0.023070,49:0.059590):0.017041, (((((50:0.028235,51:0.017075):0.005619,52:0.021828):0.032519,53:0.077006):0.009269, (54:0.002680,55:0.001144):0.046334):0.039959, (56:0.028879,57:0.037555):0.055228):0.008684):0.034212):0.021318, (58:0.085711,59:0.106281):0.012561):0.009454, ((((60:0.011286,61:0.012877):0.005152,62:0.006083):0.060188,63:0.058739):0.111665,64:0.18 4072):0.013417):0.011916, ((((((((((((65:0.011315,66:0.010853):0.010136,67:0.028098):0.001966,68:0.022315):0.012688, (69:0.015150,70:0.011028):0.013508):0.012431,71:0.040632):0.001503,72:0.046877):0.009286, (((73:0.001391,74:0.009071):0.009824, (75:0.000621,76:0.000770):0.017773):0.008570,77:0.017286):0.018761):0.068975, (78:0.042806,79:0.026245):0.067390):0.014411, ((80:0.011014,81:0.006775):0.095074,82:0.083576):0.030520):0.021160, ((((83:0.005265,84:0.009916):0.037452,85:0.057418):0.014483,86:0.056614):0.035981, ((87:0.055343,88:0.056073):0.028254,89:0.070878):0.008242):0.006244):0.004449, (((((((90:0.017250,91:0.026918):0.003821,92:0.024139):0.011103,93:0.035835):0.004701,94:0. 024822):0.013126,95:0.035799):0.024557, (96:0.015147,97:0.013557):0.057038):0.002701,98:0.084447):0.031336):0.053348, (99:0.019979,100:0.054510):0.124227):0.036319):0.033248,101:0.229015):0.039247, ((((((((((((102:0.015226,103:0.020402):0.008827,104:0.018705):0.002909, ((105:0.018282,106:0.012659):0.006255,107:0.023774):0.021767):0.002055, (108:0.023575,109:0.030901):0.001356):0.020982, ((110:0,111:0):0.027405,112:0.032483):0.028444):0.008721, ((((113:0.013158,114:0.024564):0.009625,115:0.031256):0.008533,116:0.043504):0.003283, (117:0.012319,118:0.010618):0.018186):0.012295):0.007265,119:0.049330):0.015539, ((120:0.038933,121:0.043099):0.011472,122:0.050219):0.026190):0.002966,123:0.090090):0.00 6406,124:0.052180):0.046537, (((125:0.017954,126:0.019120):0.021664,127:0.016663):0.019702,128:0.028151):0.058837):0.2 21825,(((((((129:0.017215,130:0.026818):0.007730,131:0.019798):0.015148, (132:0.005563,133:0.004239):0.031161):0,134:0.044936):0.026159,135:0.060254):0.022021, ((136:0.039763,137:0.039886):0.014560,138:0.045229):0.019272):0.074851,
8 ((139:0.048562,140:0.034418):0.015213,141:0.040739):0.103251):0.071929):0.010670):0.0199 60,(((((((142:0.020145,143:0.017967):0.013693,144:0.031524):0.011347, (145:0.026583,146:0.028139):0.015285):0.053513, (147:0.047322,148:0.042297):0.074940):0.054840,149:0.112759):0.053712, ((150:0.040765,151:0.051565):0.107354,152:0.119213):0.155898):0.047868, (((153:0.006824,154:0.015793):0.062312,155:0.045887):0.044891, (156:0.000921,157:0.022243):0.130476):0.115038):0.033836):0.121682, ((158:0.418608,159:0.302185):0.075587,(160:0.192151,161:0.213578):0.187695):0.083174, ((((162:0.157603,163:0.140732):0.332705,(164:0.002224,165:0.005336):0.258064):0.036831, (166:0.162453,167:0.093899):0.133182):0.006786,((168:0.274789,169:0.241921):0.197359, (170:0.141583,171:0.119841):0.113981):0.028500):0.017793); End;
9