Supplementary Information
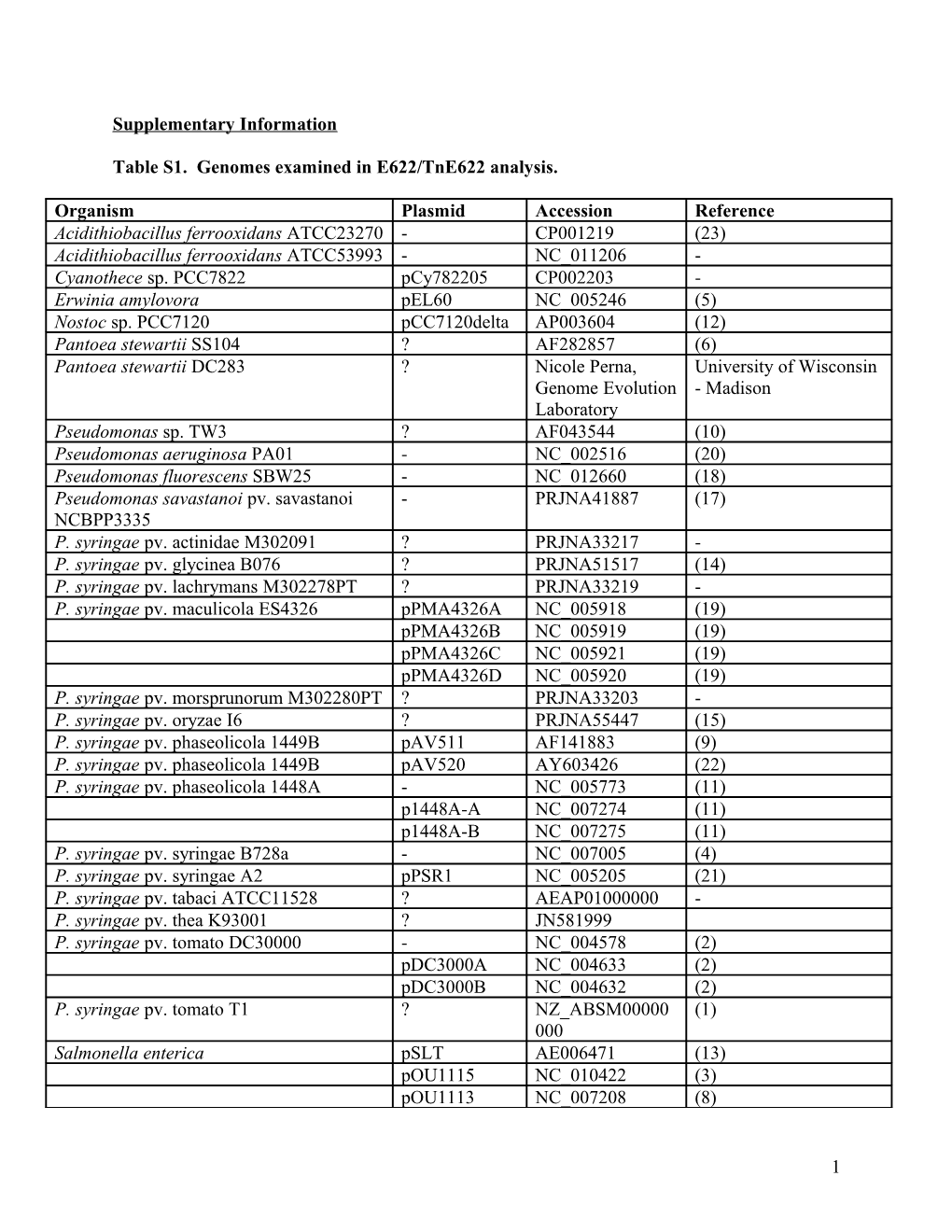
Table S1. Genomes examined in E622/TnE622 analysis.
Organism Plasmid Accession Reference Acidithiobacillus ferrooxidans ATCC23270 - CP001219 (23) Acidithiobacillus ferrooxidans ATCC53993 - NC_011206 - Cyanothece sp. PCC7822 pCy782205 CP002203 - Erwinia amylovora pEL60 NC_005246 (5) Nostoc sp. PCC7120 pCC7120delta AP003604 (12) Pantoea stewartii SS104 ? AF282857 (6) Pantoea stewartii DC283 ? Nicole Perna, University of Wisconsin Genome Evolution - Madison Laboratory Pseudomonas sp. TW3 ? AF043544 (10) Pseudomonas aeruginosa PA01 - NC_002516 (20) Pseudomonas fluorescens SBW25 - NC_012660 (18) Pseudomonas savastanoi pv. savastanoi - PRJNA41887 (17) NCBPP3335 P. syringae pv. actinidae M302091 ? PRJNA33217 - P. syringae pv. glycinea B076 ? PRJNA51517 (14) P. syringae pv. lachrymans M302278PT ? PRJNA33219 - P. syringae pv. maculicola ES4326 pPMA4326A NC_005918 (19) pPMA4326B NC_005919 (19) pPMA4326C NC_005921 (19) pPMA4326D NC_005920 (19) P. syringae pv. morsprunorum M302280PT ? PRJNA33203 - P. syringae pv. oryzae I6 ? PRJNA55447 (15) P. syringae pv. phaseolicola 1449B pAV511 AF141883 (9) P. syringae pv. phaseolicola 1449B pAV520 AY603426 (22) P. syringae pv. phaseolicola 1448A - NC_005773 (11) p1448A-A NC_007274 (11) p1448A-B NC_007275 (11) P. syringae pv. syringae B728a - NC_007005 (4) P. syringae pv. syringae A2 pPSR1 NC_005205 (21) P. syringae pv. tabaci ATCC11528 ? AEAP01000000 - P. syringae pv. thea K93001 ? JN581999 P. syringae pv. tomato DC30000 - NC_004578 (2) pDC3000A NC_004633 (2) pDC3000B NC_004632 (2) P. syringae pv. tomato T1 ? NZ_ABSM00000 (1) 000 Salmonella enterica pSLT AE006471 (13) pOU1115 NC_010422 (3) pOU1113 NC_007208 (8)
1 pSCV50 CM001063 (16) pKDSC50 NC_002638 (7) Shewanella baltica OS185 - NC_009665 - Shewanella baltica OS195 - NC_009997 - Shewanella baltica OS223 - CP001252 - Shewanella baltica OS678 - CP002383 - Yersinia pestis CO92 - AL590842
References
1. Almeida, N. F., S. Yan, M. Lindeberg, D. J. Studholme, D. J. Schneider, B. Condon, H. J. Liu, C. J. Viana, A. Warren, C. Evans, E. Kemen, D. MacLean, A. Angot, G. B. Martin, J. D. Jones, A. Collmer, J. C. Setubal, and B. A. Vinatzer. 2009. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Molecular Plant-Microbe Interactions 22:52-62. 2. Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. W. Zhou, J. Liu, Q. P. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Y. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America 100:10181-10186. 3. Chu, C. S., Y. Feng, A. C. Chien, S. N. Hu, C. H. Chu, and C. H. Chiu. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339-343. 4. Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proceedings Of The National Academy Of Sciences Of The United States Of America 102:11064-11069. 5. Foster, G. C., G. C. McGhee, A. L. Jones, and G. W. Sundin. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Applied and Environmental Microbiology 70:7539-7544. 6. Frederick, R. D., M. Ahmad, D. R. Majerczak, A. S. Arroyo-Rodriguez, S. Manulis, and D. L. Coplin. 2001. Genetic organization of the Pantoea stewartii subsp stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Molecular Plant-Microbe Interactions 14:1213-1222.
2 7. Haneda, T., N. Okada, N. Nakazawa, T. Kawakami, and H. Danbara. 2001. Complete DNA sequence and comparative analysis of the 50-kilobase virulence plasmid of Salmonella enterica serovar choleraesuis. Infection and Immunity 69:2612-2620. 8. Hong, S. F., C. H. Chiu, C. S. Chu, Y. Feng, and J. T. Ou. 2008. Complete nucleotide sequence of a virulence plasmid of Salmonella enterica serovar Dublin and its phylogenetic relationship to the virulence plasmids of serovars Choleraesuis, Enteritidis and Typhimurium. Fems Microbiology Letters 282:39-43. 9. Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma, D. L. Arnold, M. J. Gibbon, J. Murillo, J. D. Taylor, and A. Vivian. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proceedings of the National Academy of Sciences, USA 96:10875-10880. 10. James, K. D., and P. A. Williams. 1998. ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. Journal of Bacteriology 180:2043-2049. 11. Joardar, V., M. Lindeberg, R. Jackson, J. Selengut, R. Dodson, L. Brinkac, S. Daugherty, R. Deboy, A. Durkin, M. Giglio, R. Madupu, W. Nelson, M. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. Fraser, A. Chatterjee, S. Cartinhour, D. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole- genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. Journal of Bacteriology 187:6488-6498. 12. Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp strain PCC 7120. DNA Research 8:205-213. 13. McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Y. Du, S. F. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature 413:852-856. 14. Qi, M. S., D. P. Wang, C. A. Bradley, and Y. F. Zhao. 2011. Genome sequence analyses of Pseudomonas savastanoi pv. glycinea and subtractive hybridization-based comparative genomics with nine pseudomonads. Plos One 6. 15. Reinhardt, J. A., D. A. Baltrus, M. T. Nishimura, W. R. Jeck, C. D. Jones, and J. L. Dangl. 2009. de novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Research 19:294-305. 16. Richardson, E. J., B. Limaye, H. Inamdar, A. Datta, K. S. Manjari, G. D. Pullinger, N. R. Thomson, R. R. Joshi, M. Watson, and M. P. Stevens. 2011. Genome sequences of Salmonella enterica serovar typhimurium, choleraesuis, dublin, and gallinarum strains of well-defined virulence in food-producing animals. Journal of Bacteriology 193:3162- 3163.
3 17. Rodriguez-Palenzuela, P., I. M. Matas, J. Murillo, E. Lopez-Solanilla, L. Bardaji, I. Perez-Martinez, M. E. Rodriguez-Moskera, R. Penyalver, M. M. Lopez, J. M. Quesada, B. S. Biehl, N. T. Perna, J. D. Glasner, E. L. Cabot, E. Neeno-Eckwall, and C. Ramos. 2010. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environmental Microbiology 12:1604-1620. 18. Silby, M. W., A. M. Cerdeno-Tarraga, G. S. Vernikos, S. R. Giddens, R. W. Jackson, G. M. Preston, X. X. Zhang, C. D. Moon, S. M. Gehrig, S. A. C. Godfrey, C. G. Knight, J. G. Malone, Z. Robinson, A. J. Spiers, S. Harris, G. L. Challis, A. M. Yaxley, D. Harris, K. Seeger, L. Murphy, S. Rutter, R. Squares, M. A. Quail, E. Saunders, K. Mavromatis, T. S. Brettin, S. D. Bentley, J. Hothersall, E. Stephens, C. M. Thomas, J. Parkhill, S. B. Levy, P. B. Rainey, and N. R. Thomson. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biology 10. 19. Stavrinides, J., and D. S. Guttman. 2004. Nucleotide sequence and evolution of the five-plasmid complement of the phytopathogen Pseudomonas syringae pv. maculicola ES4326. Journal of Bacteriology 186:5101-5115. 20. Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. 21. Sundin, G. W., C. T. Mayfield, Y. Zhao, T. S. Gunasekera, G. L. Foster, and M. S. Ullrich. 2004. Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2. Molecular Genetics and Genomics 270:462-475. 22. Tsiamis, G., J. W. Mansfield, R. Hockenhull, R. W. Jackson, A. Sesma, E. Athanassopoulos, M. A. Bennett, C. Stevens, A. Vivian, J. D. Taylor, and J. Murillo. 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO Journal 19:3204-3214. 23. Valdes, J., I. Pedroso, R. Quatrini, R. J. Dodson, H. Tettelin, R. Blake, J. A. Eisen, and D. S. Holmes. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9.
4 Figure S1. Genomic context of E622 inverted repeats (IRs) in genomes of Acidithiobacillus ferrooxidans, Pseudomonas syringae, Yersinia pestis. Black arrowheads represent Group I E622
IRs, blue arrowheads Group II E622 IRs. Hatching indicates pseudogenes. CH, conserved hypothetical; hyp, hypothetical.
5