Supplementary Materials
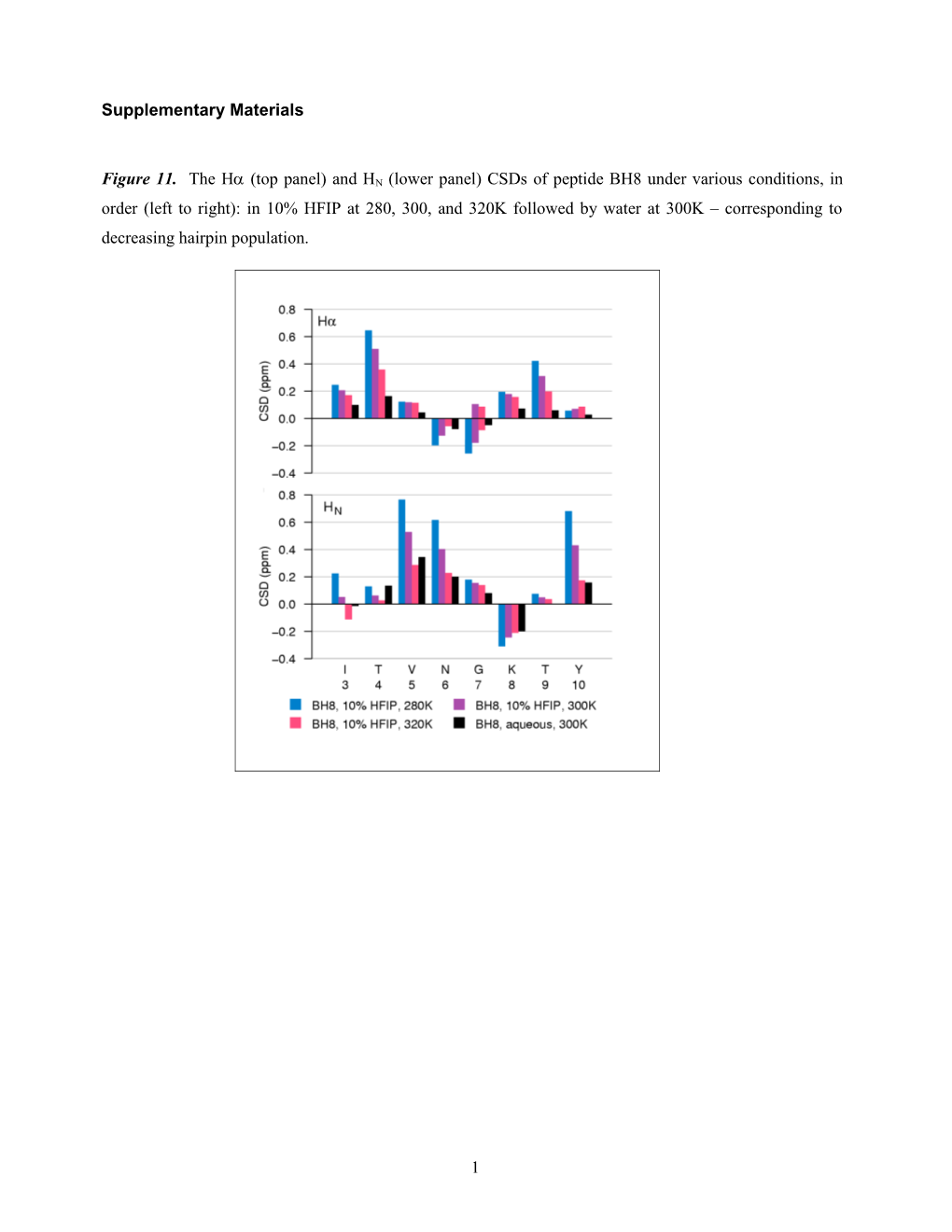
Figure 11. The H (top panel) and HN (lower panel) CSDs of peptide BH8 under various conditions, in order (left to right): in 10% HFIP at 280, 300, and 320K followed by water at 300K – corresponding to decreasing hairpin population.
1 Figure 12. H CSDs observed for ‘loop’ analogs peptide MrH3a under a variety of conditions. The [4:4]- hairpins are shown (left to right) in order of increasing fold population, all data is observations at 300K (a).
This sequence of stabilities is also apparent based on HN CSDs (Figure 6a). The [3:5]-hairpin analog, MrH6a (b). The first two data points are for this peptide at 300 K in aqueous buffer (pH 6.0) without and with the addition of HFIP to 16 vol-%. The last entry, only at strand positions, is the corresponding data for MrH3a in 8% HFIP at 300 K (the temperature at which folding is maximal).
2 To confirm the generality of the [3:5]-hairpin CSD patterns, we also examined the UTH3 sequence of deAlba et al (de Alba et al. 1999). UTH3 was examined at pH 3.2 and 5.8 (10mM phosphate aqueous buffer) and with the addition TFE to a final 30 vol-%. The CSD histograms for this sequence under conditions that favor folding to different extents are shown in Figure 13. In 30% TFE, both H and HN CSDs (notably I3, W11, V13, Y4N, T12N and T14N) approach 1 ppm, a value that represents 90+% strand alignment in the [2:2]-hairpins. The H sites with large positive CSDs are fully out-of-phase with
10 the large HN CSDs. In addition, the Thr HN is shifted upfield indicating its S+1 disposition. As in MrH6a, the Asp at T2 has an upfield shifted HN. The melting behavior of this hairpin in 30% TFE is also shown. A monotonic loss of CSD magnitude throughout the sequence, whether the CSDs are positive or negative, was observed on warming; this suggests fully cooperative melting.
Figure 13. H (upper) and HN (lower panel) CSDs observed for peptide UTH3 under a variety of conditions, in order (left to right): 30% TFE at 280, 300 and 320 K and in water at pH 5.8 and 3.2. The aqueous data is for 280K. The H sites with large positive CSDs (I3,W11 and V13) correspond to the S4, S+2 and S+4 strand positions.
3 4 The value of L-Pro substitutions for obtaining reference shifts for 3-stranded sheet analyses
A recent publication from the Gellman lab (Syud et al. 2003) continues to advocate the use of the shifts of outwardly directed H’s of specific H-bonded residue sites. To provide further insights into the use of H shifts at H-bonded sites for folding measurement and the proper referencing procedure for such applications, we have calculated fold estimates over the two hairpins following Gellman’s procedure using the two L-Pro-Gly mutants as the 0% folded reference values and taking the largest shift observed for I3 (S- 3), S5 (S1), K8 (S+1), V13 (S1), Orn16 (S+1), and I18 (S+3) as the 100% values. The I3, S5 and K8 measures provide the %-fold for the 1st hairpin; V13, O16 and I18 provide the measure for the 2nd hairpin. This should give data that is comparable to that in Table 1 where the same 100% calibration was employed; the correlation appears in Figure 14a.
It is apparent that the data from S-1 sites do not track with other measures of fold population. Even when these are removed, the correlation with fold estimates based on inwardly directed H and HN strand resonances is modest (R2 = 0.78) but the slope is near unity and the least squares line goes through the origin. There are a number of instances in which the two remaining measures for each hairpin f F differ substantially, by up to factor of 2.2 and fF = 0.40. Nonetheless, the average of the I3 and K8 measures (monitoring folding about turn 1), and the average of O16 and I18 for turn 2, do reflect fold population and provide a significantly improved correlation (R2 = 0.88), Figure 16b. Using the same measures, we examined the relative merits of assuming that 0% folding should result in CSD = 0 versus using the L-Pro mutants as the 0% references (Figure 16c). The correlation (R2 = 0.81) assuming that CSD = 0 corresponds to 0% folding is clearly inferior to that using the L-Pro-control referencing method (R2 = 0.88). This likely reflects sequence specific effects on random coil shifts that are not included in our CSD calculation algorithm. The small CSDs observed for H-bonded H sites in the L-Pro-Gly turn species (+0.042 ± 0.028) are significant since these sites display rather small structuring shifts, 0.26 ± 0.04 ppm for 100% folding. This is reflected in a y intercept of 19% folded in Figure 16c, which contrasts to the zero intercept value in Figure 16b.
5 Figure 14. Three-stranded sheet mutants: The correlation between “relative %-folded measures” derived from structuring shifts at H in H-bonded strand residues of 3-stranded sheet mutants and the prior estimates appearing in Table 1: both axes are %-folded with SG(pG-pG) in 8% HFIP set at 100%. In (a), the S-1 sites are shown in red (filled = S5, open = V13), S+1 sites in green (K8, Orn16), and S3 sites in blue (filled = I18, open = I3). The S-1 sites are excluded from the least squares linear correlation. The illustration in (b) employs average structuring shifts - I3 and K8 for the N-terminal hairpin, O16 and I18 for the C-terminal hairpin – for the correlation. Both (a) and (b) employ the SG(PG-pG) and SG(pG-PG) as reference for 0% folding about the 1st and 2nd hairpin, respectively. The third panel, (c), includes the same data presented in (b), but employs our random-coil reference values (Fesinmeyer et al. 2004, see also http://andersenlab.chem.washington.edu/CSDb) rather than the shifts observed in the respective D- Pro L-Pro ‘turn’ mutants.
6