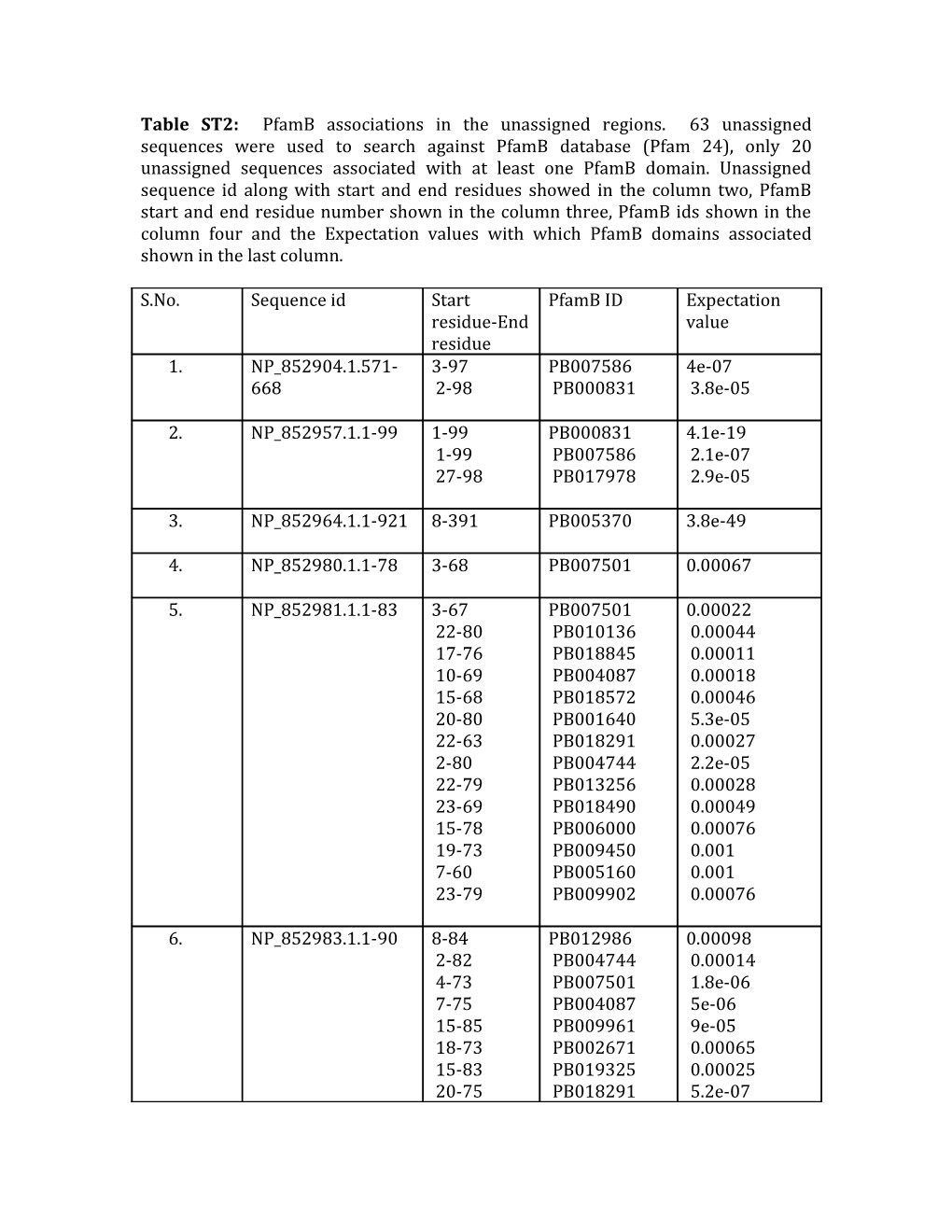
Table ST2: PfamB associations in the unassigned regions. 63 unassigned sequences were used to search against PfamB database (Pfam 24), only 20 unassigned sequences associated with at least one PfamB domain. Unassigned sequence id along with start and end residues showed in the column two, PfamB start and end residue number shown in the column three, PfamB ids shown in the column four and the Expectation values with which PfamB domains associated shown in the last column.
S.No. Sequence id Start PfamB ID Expectation residue-End value residue 1. NP_852904.1.571- 3-97 PB007586 4e-07 668 2-98 PB000831 3.8e-05
2. NP_852957.1.1-99 1-99 PB000831 4.1e-19 1-99 PB007586 2.1e-07 27-98 PB017978 2.9e-05
3. NP_852964.1.1-921 8-391 PB005370 3.8e-49
4. NP_852980.1.1-78 3-68 PB007501 0.00067
5. NP_852981.1.1-83 3-67 PB007501 0.00022 22-80 PB010136 0.00044 17-76 PB018845 0.00011 10-69 PB004087 0.00018 15-68 PB018572 0.00046 20-80 PB001640 5.3e-05 22-63 PB018291 0.00027 2-80 PB004744 2.2e-05 22-79 PB013256 0.00028 23-69 PB018490 0.00049 15-78 PB006000 0.00076 19-73 PB009450 0.001 7-60 PB005160 0.001 23-79 PB009902 0.00076
6. NP_852983.1.1-90 8-84 PB012986 0.00098 2-82 PB004744 0.00014 4-73 PB007501 1.8e-06 7-75 PB004087 5e-06 15-85 PB009961 9e-05 18-73 PB002671 0.00065 15-83 PB019325 0.00025 20-75 PB018291 5.2e-07 19-88 PB009767 7.1e-06 20-79 PB018104 0.00023 17-75 PB012999 0.00062 14-80 PB006000 0.0003 21-75 PB013256 0.00051 22-73 PB006333 0.001
7. NP_852984.1.1-99 8-92 PB012986 0.00016 16-96 PB009961 1.7e-06 11-64 PB000827 0.00011 19-97 PB009767 9.6e-08 21-79 PB018104 3.4e-05 16-93 PB019325 5.6e-05 17-75 PB018845 0.00013 18-94 PB016851 0.00068 20-76 PB018291 7.7e-09 9-96 PB004087 1.2e-07 5-65 PB005160 3.8e-06 14-92 PB006000 1.1e-05 18-95 PB009450 1.8e-05 19-73 PB002671 0.00011 17-95 PB018134 0.00019 21-75 PB013256 2.5e-05 18-96 PB003907 7.1e-05 8-97 PB005207 0.00016 21-75 PB013734 0.00029 16-99 PB018572 4.7e-05 22-91 PB001640 0.0001 23-96 PB014100 0.00061 23-98 PB006333 5.8e-06 23-69 PB018490 3.6e-05 23-97 PB009902 0.00029
8. NP_852986.1.1-80 15-78 PB009961 0.001 4-73 PB007501 0.0006 17-76 PB001640 0.00027 16-62 PB018572 0.00072 8-65 PB004087 0.00039 22-60 PB018291 0.00039 19-76 PB001024 0.00036 23-61 PB018490 0.00052
9. NP_853120.1.1-75 19-54 PB018291 0.00013
10. NP_853121.1.1-81 4-75 PB007501 0.00018 12-78 PB016763 0.00092 3-79 PB012325 0.00013 7-66 PB012999 0.00024 14-73 PB009961 0.00029 17-77 PB001640 0.0004 6-60 PB004087 0.00048 4-77 PB004744 0.00054
11. NP_853124.1.1-78 3-65 PB007501 0.00025 10-63 PB004087 0.00088 21-57 PB018291 0.00028
12. NP_853125.1.1-74 3-68 PB007501 0.00044 14-73 PB009961 0.00096 22-73 PB010136 0.00014 20-71 PB001640 0.00014 7-69 PB004087 9.6e-06 17-69 PB009767 0.00048 15-69 PB018572 6.5e-05 18-74 PB018845 0.00049 4-62 PB005160 2.1e-05 22-73 PB013256 0.00028 16-73 PB006000 0.00069 20-67 PB008447 0.00092 2-70 PB004744 3.7e-05 23-65 PB018291 0.00011 17-70 PB009450 0.00017 20-71 PB001024 0.00063 23-69 PB018490 2.6e-05 22-73 PB006333 0.00016 23-72 PB009902 0.00023 23-71 PB014100 0.00047 7-71 PB005207 0.00046
13. NP_853126.1.1-106 11-94 PB004087 2.3e-06 15-92 PB012999 2.6e-05 19-97 PB009767 1.9e-05 17-94 PB003907 0.00016 14-93 PB006000 8.9e-05 16-102 PB016763 0.00028 21-86 PB018291 0.00083 19-95 PB009450 9.8e-05 18-96 PB018134 0.00047 14-96 PB001024 8.9e-05 23-100 PB009902 9.3e-05 10-89 PB005207 0.00025
14. NP_853175.1.125- 17-105 PB002899 1.4e-06 230 15. NP_853286.1.341- 42-78 PB012318 3e-08 457 42-110 PB016297 6.2e-12 53-93 PB013215 0.00041
16. NP_853356.1.101- 2-90 PB005829 4.3e-07 192 17. NP_853371.1.224- 1-47 PB000097 1.4e-09 325 18. NP_853477.1.1-204 9-192 PB001981 1.9e-54
19. NP_853486.1.1-691 6-148 PB004087 6.6e-06 32-144 PB009450 0.00016 10-133 PB001024 5.3e-06 42-140 PB005319 0.00011 35-149 PB018285 0.00065 5-124 PB005160 2.9e-06 47-182 PB013256 1.1e-06 38-166 PB009767 2.1e-05 232-663 PB001981 4.2e-140
20. NP_853489.1.1-453 23-441 PB001981 5.7e-144