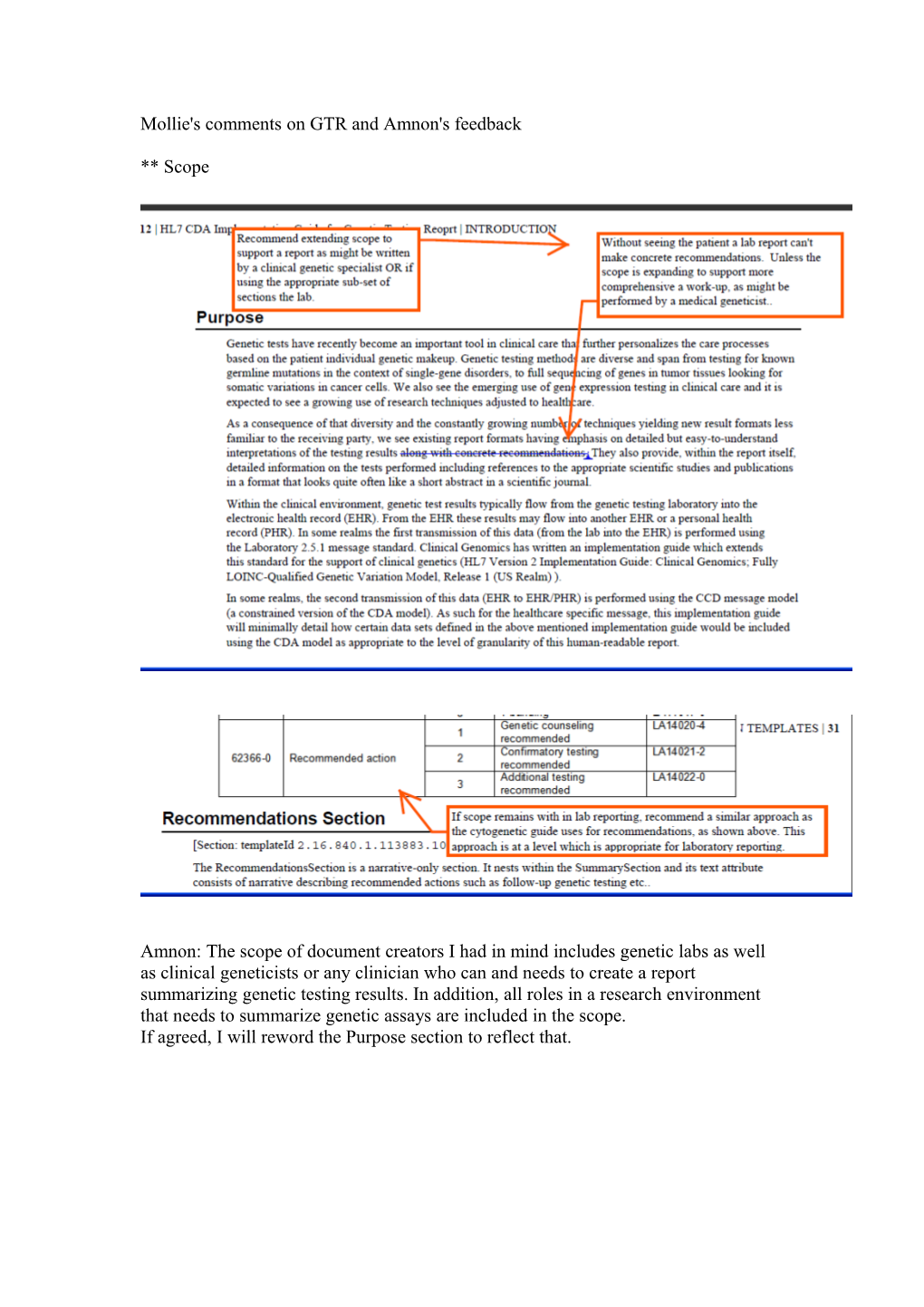
Mollie's comments on GTR and Amnon's feedback
** Scope
Amnon: The scope of document creators I had in mind includes genetic labs as well as clinical geneticists or any clinician who can and needs to create a report summarizing genetic testing results. In addition, all roles in a research environment that needs to summarize genetic assays are included in the scope. If agreed, I will reword the Purpose section to reflect that. ** Recommendations
Amnon: Since the Universal GTR specification needs to serve both clinical and research environment, how about we reword this bullet to say: Emphasize interpretations and recommendations (in the clinical environment, interpretations should be related to clinically relevant findings) ** Care Plan section
Amnon: This is a scope issue. I agree that a care plan seems somewhat out of scope for a genetic testing report, especially as we find it in CCD or the v3 Patient Care specs. So, to avoid overall, perhaps we need to take it out all together
By the way, what's the difference between 'lab recommendations' and 'recommendation'? Note that this section resides within the 'recommendation' section.
** Follow Up Visits To Specialists
Amnon: In this case I think we should leave this section in place as I noticed such section in many existing reports. Also, it's different than Care Plan because it's stemming from the testing results as assessed by the lab or geneticist. ** Other Tests
Amnon: The section is supposed to be a catcher of any test that doesn't fall into one of the specialized sections (e.g., Genetic Variation). In all sections, the section type is coded through a LOINC code (still TBD) but in this case it's obvious that we can't get a code for such a section, perhaps we can use the document type code as a general genetic analysis. As for the codifying the test findings & interpretation, it's residing in inner structures by using codes of observations etc. In general, it's encouraged to codify those structures as much as possible, and we can add that as a general recommendation of a GTR actual implementation. ** Overall Interpretation Section
Amnon: #1 comment: This is supposed to be a Section code which is part of all section codes we need to ask from LOINC.
#2 comment: We do reuse the current LOINC codes. If you follow the link to the actual observation template, you'll see that the code and value attributes are bound to the appropriate LOINC value set. ** References Section
Amnon: Great idea! I will reword the description of the section so that it isn't 'narrative-only' section, rather should consists of the references in structured entries, using pubmed, OMIM or any internationally-recognized registration of publications. ** Phenotype and Observation
Amnon: The words Phenotype and Observation in this context are part of string name that starts with Overall and ends with Analysis and thus should be understood in this context.
In addition, the main distinction we have in the Clinical Genomics Domain is between observed and interpretive phenotypes which are all observations. If the patient was examined and the phenotype is associated to the genetic observation (e.g., observed responsiveness to Gefintinib associated with EGFR mutation), then it's an observed phenotype otherwise, it's interpretive (e.g., we found this mutation and believe it could be associated with this responsiveness to Gefintinib).
In this specific example, you highlighted in yellow only the two words "Phenotype Observation", but as said, the word "Interpretive" is preceding these words and makes it an interpretation of some genomic observation. ** Repeating Genomic Source Class
Amnon: The way I designed the GTR is that the Summary Section sets some context for all other sections with regard to specific types of information such as the specimen and genomic source class. This is meant to reduce repetition if not needed, e.g., all genetic variations are observed in the same specimen and same genomic source class. Unlike messages, a CDA document has to be parsed in its entirety and extracted pieces of it might be meaningless. The best example is the header which sets the context for the entire document, e.g., you don't repeat the patient ids in each observation unless a specific observation relates to another subject, e.g., the father of the patient and then you can override the header.
However, you feel it's safer to repeat those pieces of information in each section (which implies repetition in each finding), I'm willing to change the guide accordingly. ** Constraining conventions
Amnon: The LOINC codes are reused – just follow the link above your comment and you should see the LOINC value set.
Regarding the use of 'self', this was done as part of an effort to see how we phrase OCL (Object Constraining Language) statements that constrains the CDA spec to say that if you assigned to the code attribute the code 53040-2 (LOINC code for Drug metabolism sequence variation interpretation), then the value attribute SHALL be drawn from the following LOINC answer list: Ultrarapid metabolizer=LA10315-2; Extensive metabolizer=LA10316-0; Intermediate metabolizer=LA10317-8; Poor metabolizer=LA9657-3
In this way the guide is still Universal but has explicit constraints on using the LOINC codes, for example you cannot mix and match different vocabularies by choosing a code from LOINC for the code attribute and a code from another vocabulary for the value attribute.