FRDC Final Report Design Standard
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sabine) Possesses a More Elongate Triangular, Acute
Kensley, Tranter & Griffin: Deep water decapod Crustacea 309 Wicksten & Mendez, 1982, and L. splendidus somite 6; posteroventral angle of somite 6 with small Wicksten & Mendez, 1982. tooth; posterolateral lobe overlapping telsonic base Lebbeus polaris (Sabine) possesses a more elongate triangular, acute. Telson dorsally gently convex, with rostrum than the present species, and does not have a 2 pairs dorsolateral spines in posterior half; posterior high-crested carapace. Lebbeus brandtii margin broadly triangular, with 3 pairs of spines, (Brazhnikov) has a short rostrum lacking, or with second pair longest. only one ventral tooth. Lebbeus grandimanus Cornea of eye much wider than eyestalk, well (Brazhnikov) has a relatively narrow rostrum, seen in pigmented; dorsal ocellus half fused to cornea. lateral view, with four postorbital teeth not forming a Antennular stylocerite lanceolate, with small basal crest. The two eastern Pacific species, L. scrippsi and tooth on outer margin, reaching distal margin of basal L. splendidus Wicksten & Mendez both have narrow antennular peduncle article; small tooth on non-crested rostra with few dorsal teeth. ventromesial margin at about distal two-thirds; Lebbeus compressus Holthuis, 1947 (= second and third peduncular articles unarmed; dorsal Spirontocaris gibberosa Yokoya, 1933), a species flagellum subequal to carapace and rostrum in length; possessing a toothed and crested carapace, has ventral flagellum somewhat longer. epipods on pereopod 1 only. (The holotypic male of Antennal scaphocerite with outer margin this species, from 232 m at Siwoya-Zaki, Japan, was straight, distal spine not reaching rounded apex of examined. Having a carapace length of 4.8 mm, the blade; basal segment with small ventrodistal tooth; specimen probably dried out at some stage, and the blade over-reaching antennular peduncle by about carapace was damaged. -

MHYC Cruising Division Program 2014 – 15 December 12Th Club Christmas Party Friday 12Th 6:30Pm (Replaces December Meeting) January 19Th End of Cruise BBQ
Volume No. 34, No. 11 December 2014 Editor Michael Mulholland-Licht Next Meeting: FRIDAY DECEMBER 12 FROM 6:30PM CLUB CHRISTMAS PARTY IS IN LEU OF MEMBERS MEETING IN DECEMBER. PLEASE BOOK WITH THE OFFICE Admiral Astrid helms Bliss through the Heads on 090 exercise. Nashira (abeam) helmed by Admiral Kelly 1 CRUISING DIVISION OFFICE BEARERS – 2014 - 2015 Cruising Captain Colin Pitstock 0407-669-322 Cruising Vice Captain Phil Darling 0411-882-760 Vice-Commodore Colin Pitstock 0407-669-322 Cruising Secretary Michael Mulholland-Licht 0418 476 216 Treasurer Trevor D’Alton 9960-2878 Membership Jean Parker 0403-007-675 Name Tags Lena D’Alton 9960-2878 Compass Rose Maralyn Miller and Committee Members 0411-156-009 Coordinator Safety Coordinator Bill Allen 9977- 0392 Waterways User Group Mike McEvoy 9968-1777 Sailing Committee Colin Pitstock 0407-669-322 Guest Speakers Royce Englehardt, & Committee Members as required On Water Events Colin Pitstock/ Michael Mulholland- Michael 0418-476-216 Coordinator Licht/ Phil Darling Phil 0411-882-760 On Land Events Jean Parker / Hilary Gallagher Coordinators General Committee Royce Englehardt, Trevor D’Alton, Phil Darling, Maralyn Miller, assistance Hilary Gallagher / Paul Wotherspoon Editor's note: Deadline for the next edition of the Compass Rose, is: 1st February 2015 The EDITOR for the next Compass Rose is Trevor D’Alton. Please forward contributions c/-: MHYC PO Box 106 SEAFORTH NSW 2092, Or Email: [email protected] Opinions expressed in the Compass Rose are those of the contributors, and do not necessarily reflect opinions of either Middle Harbour Yacht Club or the Cruising Division 2 MHYC Cruising Division Program 2014 – 15 December 12th Club Christmas Party Friday 12th 6:30pm (Replaces December meeting) January 19th End of Cruise BBQ. -

Commercial-In-Confidence
Any use of the Report, use of any part of it, or use of the names NewSouth Global, Expert Opinion Services, University of New South Wales, UNSW, the name of any unit of the University or the name of the Consultant, in direct or in indirect advertising or publicity, is forbidden. COMMERCIAL-IN-CONFIDENCE Report prepared on behalf of NSG Consulting A division of NewSouth Global Pty Limited Ecological issues in relation to BlueScope Steel SCP proposed salt water cooling for CH2M HILL Australia Pty Ltd by Dr Emma Johnston, Dr Jan Carey and Dr Nathan Knott August 2006 J069413 The University of New South Wales, Sydney 2052, DX 957 Sydney Ph: 1800 676 948 Fax: 1800 241 367 www.eos.unsw.edu.au Email: [email protected] CONTENTS Page Executive Summary: ................................................................................................ 1 Introduction............................................................................................................... 3 Predicted Changes in Temperature ........................................................................ 4 Temperature tolerances and preferences of organisms currently found in Port Kembla Harbour........................................................................................................ 9 General temperature effects on major biochemical processes...........................14 Species expected in a slightly to moderately disturbed estuarine system........15 Aspects of current environment that may be excluding species........................17 A review of the influences -

New South Wales Records 2021 NSW State Spearfishing Records Flinders Reef S.E.Queensland
New South Wales Records 2021 NSW State Spearfishing Records Flinders Reef S.E.Queensland ANGELFISH Common Name Scientific Name Division Weight Diver Club Date Location ANGELFISH Pomacanthus semicirculatus Junior Blue Ladies Open NSW 1.020 kg S. Isles BDSC 27/12/1970 Juan & Julia Rocks Australian 3.033 kg R. Jenkinson QLD 1/6/1969 Flinders Reef S.E.Queensland BARRACUDA & PIKE Common Name Scientific Name Division Weight Diver Club Date Location BARRACUDA Syphyraena qenie Junior Blackfin Ladies Open NSW 17.660 kg E. Leeson SSD 13/03/2015 South West Rocks Australian 29.200 kg T. Neilsen UAC 10/5/2006 Cape Moreton Common Name Scientific Name Division Weight Diver Club Date Location BARRACUDA Sphyraena barracuda Junior Giant Ladies 0.865 kg J. Budworth TGCF 4/2/2018 Tweed River Open NSW 21.500 kg E. Bova SSD 18/03/2007 Coffs Harbour Australian 28.850 kg B. Paxman WA 16/4/1993 Dorre Island WA Common Name Scientific Name Division Weight Diver Club Date Location BARRACUDA Sphyraena jello Junior 8.650 kg A. Puckeridge SSD 29/03/2014 North Solitary Island Pickhandle pike Ladies Open NSW 12.927 kg P. Iredale KSC 21/02/1971 Wreck Bay Australian 15.250 kg J. Croton Qld 12/6/2015 Cape Moreton Common Name Scientific Name Division Weight Diver Club Date Location BARRACUDA Sphyraena obtusata Junior 1.700 kg A.Puckeridge SSD 7/1/2013 Coffs Harbour Striped sea pike, Ladies Open NSW 1.700 kg A.Puckeridge SSD 7/1/2013 Nth Solitary Is Australian 1.191 kg K. Wardrop NSW 28/5/1967 Whale beach, NSW Common Name Scientific Name Division Weight Diver Club Date Location BARRACOUTA Thrysites atun Junior Snoek Ladies Open NSW 0.625 kg A. -

Issues Paper for the Grey Nurse Shark (Carcharias Taurus)
Issues Paper for the Grey Nurse Shark (Carcharias taurus) 2014 The recovery plan linked to this issues paper is obtainable from: http://www.environment.gov.au/resource/recovery-plan-grey-nurse-shark-carcharias-taurus © Commonwealth of Australia 2014 This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968, all other rights are reserved. Requests and enquiries concerning reproduction and rights should be addressed to Department of the Environment, Public Affairs, GPO Box 787 Canberra ACT 2601 or email [email protected]. Disclaimer While reasonable efforts have been made to ensure that the contents of this publication are factually correct, the Commonwealth does not accept responsibility for the accuracy or completeness of the contents, and shall not be liable for any loss or damage that may be occasioned directly or indirectly through the use of, or reliance on, the contents of this publication. Cover images by Justin Gilligan Photography Contents List of figures ii List of tables ii Abbreviations ii 1 Summary 1 2 Introduction 2 2.1 Purpose 2 2.2 Objectives 2 2.3 Scope 3 2.4 Sources of information 3 2.5 Recovery planning process 3 3 Biology and ecology 4 3.1 Species description 4 3.2 Life history 4 3.3 Diet 5 3.4 Distribution 5 3.5 Aggregation sites 8 3.6 Localised movements at aggregation sites 10 3.7 Migratory movements -
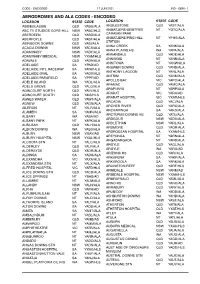
Aerodromes and Ala Codes
CODE - ENCODED 17 JUN 2021 IND - GEN - 1 AERODROMES AND ALA CODES - ENCODED LOCATION STATE CODE LOCATION STATE CODE ABBIEGLASSIE QLD YABG/ALA ANGLESTONE QLD YAST/ALA ABC TV STUDIOS GORE HILL NSW YABC/HLS ANMATJERE/GEMTREE NT YGTC/ALA ABERDEEN QLD YABD/ALA CARAVAN PARK ABERFOYLE QLD YABF/ALA ANMATJERE/PINE HILL NT YPHS/ALA STATION ABINGDON DOWNS QLD YABI/ALA ANNA CREEK SA YANK/ALA ACACIA DOWNS NSW YACS/ALA ANNA PLAINS HS WA YAPA/ALA ADAMINABY NSW YADY/ALA ANNANDALE QLD YADE/ALA ADAMINABY MEDICAL NSW YXAM/HLS ANNINGIE NT YANN/ALA ADAVALE QLD YADA/ALA ANNITOWA NT YANW/ALA ADELAIDE SA YPAD/AD ANSWER DOWNS QLD YAND/ALA ADELAIDE INTL RACEWAY SA YAIW/HLS ANTHONY LAGOON NT YANL/ALA ADELAIDE OVAL SA YAOV/HLS ANTRIM QLD YANM/ALA ADELAIDE/PARAFIELD SA YPPF/AD APOLLO BAY VIC YAPO/ALA ADELE ISLAND WA YADL/ALA ARAMAC QLD YAMC/ALA ADELS GROVE QLD YALG/ALA ARAPUNYA NT YARP/ALA AGINCOURT NORTH QLD YAIN/HLS ARARAT VIC YARA/AD AGINCOURT SOUTH QLD YAIS/HLS ARARAT HOSPITAL VIC YXAR/HLS AGNES WATER QLD YAWT/ALA ARCADIA QLD YACI/ALA AGNEW QLD YAGN/ALA ARCHER RIVER QLD YARC/ALA AILERON NT YALR/ALA ARCKARINGA SA YAKG/ALA ALAMEIN SA YAMN/ALA ARCTURUS DOWNS HS QLD YATU/ALA ALBANY WA YABA/AD ARDGOUR NSW YADU/ALA ALBANY PARK NT YAPK/ALA ARDLETHAN NSW YARL/ALA ALBILBAH QLD YALH/ALA ARDMORE QLD YAOR/ALA ALBION DOWNS WA YABS/ALA ARDROSSAN HOSPITAL SA YXAN/HLS ALBURY NSW YMAY/AD AREYONGA NT YARN/ALA ALBURY HOSPITAL NSW YXAL/HLS ARGADARGADA NT YARD/ALA ALCOOTA STN NT YALC/ALA ARGYLE QLD YAGL/ALA ALDERLEY QLD YALY/ALA ARGYLE WA YARG/AD ALDERSYDE QLD YADR/ALA ARIZONA HS -

Discussion Paper for Grey Nurse Shark Protection
Summary of Submissions Report Discussion Paper for Grey Nurse Shark Protection January 2012 1 Executive Summary • This report provides an overview of the public exhibition process and comments received in written submissions on a Discussion Paper for Grey Nurse Shark (GNS) Protection released on 31 May 2011. • The discussion paper was on public exhibition for 3 months ending 26 August 2011. • Submissions were received and analysed for comments and suggestions related to changes that could be introduced to improve the current level of protection for GNS. • A total of 960 individual submissions, 1247 form submissions (from 7 groups) and 122 petition signatures were received (2329 in total). Form submissions were treated as single submissions for the purpose of the analysis i.e. as 7 submissions. Similarly, petition signatures were treated as a single submission. However, additional individual comments received on form submissions were included in the analysis. • The most commonly raised comments and suggestions related to GNS management in submissions were: The GNS population is depleted and requires increased protection. Increase protection at Fish Rock and Green Island. All forms of fishing should be prohibited at GNS aggregation sites. Not convinced/don’t believe GNS numbers reported. More research is required about GNS population including numbers outside reported aggregation sites. Comments that individuals had observed GNS with fishing related injuries. Implement 1500 m no take sanctuary zones around all GNS aggregation sites. Reinstate closures revoked in April 2011. Some form of fishing restrictions need to be put in place at GNS aggregation sites. Increase community education programs. Comments from individuals that they had never hooked a GNS. -

Terrestrial and Marine Protected Areas in Australia
TERRESTRIAL AND MARINE PROTECTED AREAS IN AUSTRALIA 2002 SUMMARY STATISTICS FROM THE COLLABORATIVE AUSTRALIAN PROTECTED AREAS DATABASE (CAPAD) Department of the Environment and Heritage, 2003 Published by: Department of the Environment and Heritage, Canberra. Citation: Environment Australia, 2003. Terrestrial and Marine Protected Areas in Australia: 2002 Summary Statistics from the Collaborative Australian Protected Areas Database (CAPAD), The Department of Environment and Heritage, Canberra. This work is copyright. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced by any process without prior written permission from Department of the Environment and Heritage. Requests and inquiries concerning reproduction and rights should be addressed to: Assistant Secretary Parks Australia South Department of the Environment and Heritage GPO Box 787 Canberra ACT 2601. The views and opinions expressed in this document are not necessarily those of the Commonwealth of Australia, the Minister for Environment and Heritage, or the Director of National Parks. Copies of this publication are available from: National Reserve System National Reserve System Section Department of the Environment and Heritage GPO Box 787 Canberra ACT 2601 or online at http://www.deh.gov.au/parks/nrs/capad/index.html For further information: Phone: (02) 6274 1111 Acknowledgments: The editors would like to thank all those officers from State, Territory and Commonwealth agencies who assisted to help compile and action our requests for information and help. This assistance is highly appreciated and without it and the cooperation and help of policy, program and GIS staff from all agencies this publication would not have been possible. An additional huge thank you to Jason Passioura (ERIN, Department of the Environment and Heritage) for his assistance through the whole compilation process. -

Patterns of Coral Community Structure of Subtropical Reefs in the Solitary Islands Marine Reserve, Eastern Australia
MARINE ECOLOGY PROGRESS SERIES Vol. 109: 67-76.1994 Published June 9 Mar. Ecol. Prog. Ser. l Patterns of coral community structure of subtropical reefs in the Solitary Islands Marine Reserve, Eastern Australia 'Centre for Coastal Management, Southern Cross University, PO Box 157, Lisrnore, NSW 2480, Australia 2Zoology Department, University of New England, Armidale, NSW 2350, Australia ABSTRACT Although the Solitary Islands Manne Reserve lies at latitude 30" S on the east coast of Austral~a,over 700 km south of the Great Barrier Reef it contains benth~ccommunities dominated by extenslve areas of scleractln~ancorals A qual~tativesurvey publ~shedIn 1974 reported a total of 34 coral specles in the region and more recent records Include a total of 55 coral species Here, we pre- sent the results of the first quantitative benth~csurveys for 7 s~tesin the Solitary Islands Manne Reserve As a result of these surveys an additIona135 specles of scleractin~ancoral have been recorded from the region bnnging the total to 90 coral specles In 28 genera from l1 families However 21 of the 55 coral specles previously recorded were not found dunng this study These results indicate that a dynam~ctemporal pattern oi specles recultment and replacement is occurnng within these subtropical coral conununities Scleractinian coral cover ranged from a low of 8 5% at Muttonbird Island the reef closest to the coastline to 50 9% at SW Sol~taryIsland These values are wthin the range of coral cover reported for troplcal fnnging reefs Mult~d~mens~onalscal~ng (MDS) analysis -

Australian Notices to Mariners Are the Authority for Correcting Australian Charts and Publications
5 June 2009 Edition 11 Australian Notices to Mariners are the authority for correcting Australian Charts and Publications AUSTRALIAN NOTICES TO MARINERS Notices 637 - 688 Published fortnightly by the Australian Hydrographic Service Commodore R. NAIRN RAN Hydrographer of Australia SECTIONS. I. Australian Notices to Mariners, including blocks and notes. II. Hydrographic Reports. III. Navigational Warnings. For Notices affecting Admiralty List of Lights and Fog Signals; List of Radio Signals; and Sailing Directions see British Notices to Mariners. The substance of these notices should be inserted on the charts affected. Bearings are referred to the true compass and are reckoned clockwise from North; those relating to lights are given as seen by an observer from seaward. Positions quoted in permanent notices relate to the horizontal datum for the chart(s). When preliminary or temporary notices affect multiple charts, positions will be provided in relation to only one horizontal datum and that datum will be specified. When the multiple charts do not have a common horizontal datum, mariners will be required to adjust the position(s) for those charts not on the specified datum. The range quoted for a light is its nominal range. Depths are with reference to the chart datum of each chart. Heights are above mean high water springs or mean higher high water, as appropriate. The capital letter (P) or (T) after the number of any notice denotes a preliminary or temporary notice respectively, which are contained separately at the end of the permanent notices. A star (*) adjacent to the number of a notice indicates that the notice is based on original information. -

Geomorphic Features of the Continental Margin of Australia
GEOSCIENCE AUSTRALIA Geomorphic Features of the Continental Margin of Australia Peter Harris, Andrew Heap, Vicki Passlow, Laura Sbaffi, Melissa Fellows, Rick Porter-Smith, Cameron Buchanan and James Daniell Record 2003/30 SPATIAL INFORMATION FOR THE NATION Geoscience Australia Record 2003/30 Geomorphic Features of the Continental Margin of Australia Report to the National Oceans Office on the production of a consistent, high-quality bathymetric data grid and definition and description of geomorphic units for part of Australia’s marine jurisdiction Seabed Mapping and Characterisation Project Team: Peter Harris, Andrew Heap, Vicki Passlow, Laura Sbaffi, Melissa Fellows, Rick Porter-Smith, Cameron Buchanan and James Daniell Petroleum and Marine Division Geoscience Australia GPO Box 378 Canberra, ACT 2601 Department of Industry, Tourism & Resources Minister for Industry, Tourism & Resources: Senator The Hon. Ian Macfarlane, MP Parliamentary Secretary: The Hon. Warren Entsch, MP Secretary: Mark Paterson Geoscience Australia Chief Executive Officer: Dr Neil Williams © Commonwealth of Australia 2005 This work is copyright. Apart from any fair dealings for the purpose of study, research, criticism or review, as permitted under the Copyright Act 1968, no part may be reproduced by any process without written permission. Copyright is the responsibility of the Chief Executive Officer, Geoscience Australia. Requests and enquiries should be directed to the Chief Executive Officer, Geoscience Australia, GPO Box 378 Canberra ACT 2601. ISSN: 1448 - 2177 ISBN: 0 642 46791 9 GeoCat No. 61007 Bibliographic reference: Harris, P., Heap, A., Passlow, V., Sbaffi, L. Fellows, M., Porter-Smith, R., Buchanan, C., & Daniell, J. (2005). Geomorphic Features of the Continental Margin of Australia. Geoscience Australia, Record 2003/30, 142pp. -

Beyond Capricornia: Tropical Sea Slugs (Gastropoda, Heterobranchia) Extend Their Distributions Into the Tasman Sea
diversity Article Beyond Capricornia: Tropical Sea Slugs (Gastropoda, Heterobranchia) Extend Their Distributions into the Tasman Sea Matt J. Nimbs 1,2,* ID and Stephen D. A. Smith 1,2 ID 1 National Marine Science Centre, Southern Cross University, Bay Drive, Coffs Harbour, NSW 2450, Australia; [email protected] 2 Marine Ecology Research Centre, Southern Cross University, Lismore, NSW 2458, Australia * Correspondence: [email protected] Received: 7 August 2018; Accepted: 28 August 2018; Published: 4 September 2018 Abstract: There is increasing evidence of poleward migration of a broad range of taxa under the influence of a warming ocean. However, patchy research effort, the lack of pre-existing baseline data, and taxonomic uncertainty for some taxa means that unambiguous interpretation of observations is often difficult. Here, we propose that heterobranch sea slugs provide a useful target group for monitoring shifts in distribution. As many sea slugs are highly colourful, popular with underwater photographers and rock-pool ramblers, and found in accessible habitats, they provide an ideal target for citizen scientist programs, such as the Sea Slug Census. This maximises our ability to rapidly gain usable diversity and distributional data. Here, we review records of recent range extensions by tropical species into the subtropical and temperate waters of eastern Australia and document, for the first time in Australian waters, observations of three tropical species of sea slug as well as range extensions for a further six to various locations in the Tasman Sea. Keywords: range extension; climate change; heterobranch; citizen science; Sea Slug Census; biodiversity 1. Introduction By far the majority of Indo-Pacific sea slug species are tropical [1] with diversity declining away from the equator.