Ation Associated Changes in CTCF-Chromatin Binding and Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Influencers on Thyroid Cancer Onset: Molecular Genetic Basis
G C A T T A C G G C A T genes Review Influencers on Thyroid Cancer Onset: Molecular Genetic Basis Berta Luzón-Toro 1,2, Raquel María Fernández 1,2, Leticia Villalba-Benito 1,2, Ana Torroglosa 1,2, Guillermo Antiñolo 1,2 and Salud Borrego 1,2,* 1 Department of Maternofetal Medicine, Genetics and Reproduction, Institute of Biomedicine of Seville (IBIS), University Hospital Virgen del Rocío/CSIC/University of Seville, 41013 Seville, Spain; [email protected] (B.L.-T.); [email protected] (R.M.F.); [email protected] (L.V.-B.); [email protected] (A.T.); [email protected] (G.A.) 2 Centre for Biomedical Network Research on Rare Diseases (CIBERER), 41013 Seville, Spain * Correspondence: [email protected]; Tel.: +34-955-012641 Received: 3 September 2019; Accepted: 6 November 2019; Published: 8 November 2019 Abstract: Thyroid cancer, a cancerous tumor or growth located within the thyroid gland, is the most common endocrine cancer. It is one of the few cancers whereby incidence rates have increased in recent years. It occurs in all age groups, from children through to seniors. Most studies are focused on dissecting its genetic basis, since our current knowledge of the genetic background of the different forms of thyroid cancer is far from complete, which poses a challenge for diagnosis and prognosis of the disease. In this review, we describe prevailing advances and update our understanding of the molecular genetics of thyroid cancer, focusing on the main genes related with the pathology, including the different noncoding RNAs associated with the disease. -

Genotyping of Breech Flystrike Resource – Update
Project No: ON-00515 Contract No: PO4500010753 AWI Project Manager: Bridget Peachey Contractor Name: CSIRO Agriculture and Food Prepared by: Dr Sonja Dominik Publication Date: July 2019 Genotyping of breech flystrike resource – update Published by Australian Wool Innovation Limited, Level 6, 68 Harrington Street, THE ROCKS, NSW, 2000 This publication should only be used as a general aid and is not a substitute for specific advice. To the extent permitted by law, we exclude all liability for loss or damage arising from the use of the information in this publication. AWI invests in research, development, innovation and marketing activities along the global supply chain for Australian wool. AWI is grateful for its funding, which is primarily provided by Australian woolgrowers through a wool levy and by the Australian Government which provides a matching contribution for eligible R&D activities © 2019 Australian Wool Innovation Ltd. All rights reserved. Contents Executive Summary .................................................................................................................... 3 1 Introduction/Hypothesis .................................................................................................... 5 2 Literature Review ............................................................................................................... 6 3 Project Objectives .............................................................................................................. 8 4 Success in Achieving Objectives ........................................................................................ -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Extensive Variation Between Inbred Mouse Strains Due to Endogenous L1 Retrotransposition
Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition Keiko Akagi,1,5 Jingfeng Li,2,5 Robert M. Stephens,3,5 Natalia Volfovsky,3 and David E. Symer2,4,6 1Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, USA; 2Basic Research Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, USA; 3Advanced Biomedical Computing Center, Advanced Technology Program, SAIC-Frederick, Inc., Frederick, Maryland 21702, USA; 4Laboratory of Biochemistry and Molecular Biology, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, USA Numerous inbred mouse strains comprise models for human diseases and diversity, but the molecular differences between them are mostly unknown. Several mammalian genomes have been assembled, providing a framework for identifying structural variations. To identify variants between inbred mouse strains at a single nucleotide resolution, we aligned 26 million individual sequence traces from four laboratory mouse strains to the C57BL/6J reference genome. We discovered and analyzed over 10,000 intermediate-length genomic variants (from 100 nucleotides to 10 kilobases), distinguishing these strains from the C57BL/6J reference. Approximately 85% of such variants are due to recent mobilization of endogenous retrotransposons, predominantly L1 elements, greatly exceeding that reported in humans. Many genes’ structures and expression are altered directly by polymorphic L1 retrotransposons, including Drosha (also called Rnasen), Parp8, Scn1a, Arhgap15, and others, including novel genes. L1 polymorphisms are distributed nonrandomly across the genome, as they are excluded significantly from the X chromosome and from genes associated with the cell cycle, but are enriched in receptor genes. -

A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection
CLINICAL RESEARCH www.jasn.org A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection Weijia Zhang,1 Zhengzi Yi,1 Karen L. Keung,2 Huimin Shang,3 Chengguo Wei,1 Paolo Cravedi,1 Zeguo Sun,1 Caixia Xi,1 Christopher Woytovich,1 Samira Farouk,1 Weiqing Huang,1 Khadija Banu,1 Lorenzo Gallon,4 Ciara N. Magee,5 Nader Najafian,5 Milagros Samaniego,6 Arjang Djamali ,7 Stephen I. Alexander,2 Ivy A. Rosales,8 Rex Neal Smith,8 Jenny Xiang,3 Evelyne Lerut,9 Dirk Kuypers,10,11 Maarten Naesens ,10,11 Philip J. O’Connell,2 Robert Colvin,8 Madhav C. Menon,1 and Barbara Murphy1 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background In kidney transplant recipients, surveillance biopsies can reveal, despite stable graft function, histologic features of acute rejection and borderline changes that are associated with undesirable graft outcomes. Noninvasive biomarkers of subclinical acute rejection are needed to avoid the risks and costs associated with repeated biopsies. Methods We examined subclinical histologic and functional changes in kidney transplant recipients from the prospective Genomics of Chronic Allograft Rejection (GoCAR) study who underwent surveillance biopsies over 2 years, identifying those with subclinical or borderline acute cellular rejection (ACR) at 3 months (ACR-3) post-transplant. We performed RNA sequencing on whole blood collected from 88 indi- viduals at the time of 3-month surveillance biopsy to identify transcripts associated with ACR-3, developed a novel sequencing-based targeted expression assay, and validated this gene signature in an independent cohort. -

The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia
cancers Article The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia Jonathan Lukas Lühmann 1,† , Marie Stelter 1,†, Marie Wolter 1, Josephine Kater 1, Jana Lentes 1, Anke Katharina Bergmann 1, Maximilian Schieck 1 , Gudrun Göhring 1, Anja Möricke 2, Gunnar Cario 2, Markéta Žaliová 3 , Martin Schrappe 2, Brigitte Schlegelberger 1, Martin Stanulla 4 and Doris Steinemann 1,* 1 Department of Human Genetics, Hannover Medical School, 30625 Hannover, Germany; [email protected] (J.L.L.); [email protected] (M.S.); [email protected] (M.W.); [email protected] (J.K.); [email protected] (J.L.); [email protected] (A.K.B.); [email protected] (M.S.); [email protected] (G.G.); [email protected] (B.S.) 2 Department of Pediatrics I, ALL-BFM Study Group, Christian-Albrechts University Kiel and University Medical Center Schleswig-Holstein, 24105 Kiel, Germany; [email protected] (A.M.); [email protected] (G.C.); [email protected] (M.S.) 3 Department of Paediatric Haematology and Oncology, 2nd Faculty of Medicine, Charles University and University Hospital Motol, CZ-15006 Prague, Czech Republic; [email protected] 4 Pediatric Hematology and Oncology, Hannover Medical School, 30625 Hannover, Germany; [email protected] * Correspondence: [email protected] † These authors equally contributed to this work. Citation: Lühmann, J.L.; Stelter, M.; Wolter, M.; Kater, J.; Lentes, J.; Bergmann, A.K.; Schieck, M.; Simple Summary: The stratification of childhood ALL is currently based on various diagnostic Göhring, G.; Möricke, A.; Cario, G.; assays. -

GOLGA5 Purified Maxpab Rabbit Polyclonal Antibody (D01P)
GOLGA5 purified MaxPab rabbit polyclonal antibody (D01P) Catalog # : H00009950-D01P 規格 : [ 100 ug ] List All Specification Application Image Product Rabbit polyclonal antibody raised against a full-length human GOLGA5 Western Blot (Tissue lysate) Description: protein. Immunogen: GOLGA5 (NP_005104.2, 1 a.a. ~ 731 a.a) full-length human protein. Sequence: MSWFVDLAGKAEDLLNRVDQGAATALSRKDNASNIYSKNTDYTELHQQN TDLIYQTGPKSTYISSAADNIRNQKATILAGTANVKVGSRTPVEASHPVEN enlarge ASVPRPSSHFVRRKKSEPDDELLFDFLNSSQKEPTGRVEIRKEKGKTPV FQSSQTSSVSSVNPSVTTIKTIEENSFGSQTHEAASNSDSSHEGQEESSK Western Blot (Tissue lysate) ENVSSNAACPDHTPTPNDDGKSHELSNLRLENQLLRNEVQSLNQEMASL LQRSKETQEELNKARARVEKWNADHSKSDRMTRGLRAQVDDLTEAVAA KDSQLAVLKVRLQEADQLLSTRTEALEALQSEKSRIMQDQSEGNSLQNQ ALQTFQERLHEADATLKREQESYKQMQSEFAARLNKVEMERQNLAEAIT LAERKYSDEKKRVDELQQQVKLYKLNLESSKQELIDYKQKATRILQSKEK LINSLKEGSGFEGLDSSTASSMELEELRHEKEMQREEIQKLMGQIHQLRS ELQDMEAQQVNEAESAREQLQDLHDQIAGQKASKQELETELERLKQEF HYIEEDLYRTKNTLQSRIKDRDEEIQKLRNQLTNKTLSNSSQSELENRLHQ enlarge LTETLIQKQTMLESLSTEKNSLVFQLERLEQQMNSASGSSSNGSSINMSG IDNGEGTRLRNVPVLFNDTETNLAGMYGKVRKAASSIDQFSIRLGIFLRRY Western Blot (Cell lysate) PIARVFVIIYMALLHLWVMIVLLTYTPEMHHDQPYGK Host: Rabbit Reactivity: Human, Mouse enlarge Quality Control Antibody reactive against mammalian transfected lysate. Testing: Western Blot (Transfected lysate) Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download enlarge Datasheet: Download Immunofluorescence Applications Western Blot (Tissue lysate) -
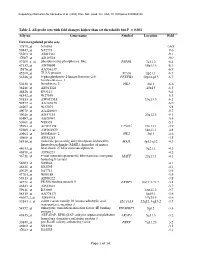
Table 2. All Probe Sets with Fold Changes Higher Than Set Thresholds but P > 0.001 Affy No
Supporting information for Sanoudou et al. (2003) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0330960100 Table 2. All probe sets with fold changes higher than set thresholds but P > 0.001 Affy no. Gene name Symbol Location Fold Down-regulated probe sets 47879_at N46863 -16.9 59447_at N52773 -9.6 55503_at AI085361 -9.1 47087_at AI310524 -9.0 37209_g_at phosphoserine phosphatase-like PSPHL 7q11.2 -8.4 47137_at AI479899 19p13.3 -8.1 45876_at AA536137 -6.9 45260_at TU3A protein TU3A 3p21.1 -6.7 56246_at 6-phosphofructo-2-kinase/fructose-2,6- PFKFB3 10p14-p15 -6.7 bisphosphatase 3 50230_at hexokinase 2 HK2 2p13 -6.6 38248_at AB011124 20p13 -6.5 44426_at R93141 -6.4 48542_at W27559 -6.3 53813_at AW051518 13q13.3 -6.1 59577_at AA243670 -6.0 46907_at W37075 -5.8 49078_at AA424983 -5.7 49026_at AI357153 20p12.3 -5.4 53487_at AI670947 -5.4 50161_at N39328 -5.1 35994_at AC002398 F25965 19q13.1 -4.9 52285_f_at AW002970 18p11.1 -4.8 40964_at hexokinase 2 HK2 2p13 -4.6 49806_at AI932283 -4.5 58918_at molecule possessing ankyrin repeats induced by MAIL 3p12-q12 -4.3 lipopolysaccharide (MAIL), homolog of mouse 44633_at heat shock 27 kDa associated protein 3q21.1 -4.3 46858_at AI796221 -4.2 36711_at v-maf musculoaponeurotic fibrosarcoma oncogene MAFF 22q13.1 -4.1 homolog F (avian) 54683_at N49844 -4.1 46621_at N32595 -4.1 49629_at N47713 -3.9 47703_at W89189 -3.9 59313_at AI598222 -3.8 34721_at FK506 binding protein 5 FKBP5 6p21.3-21.2 -3.8 46843_at AI632621 -3.7 59611_at R53069 16p11.2 -3.7 58315_at AA778171 3p25.1 -3.6 46607_f_at AI885018 17q25.3 -3.6 33143_s_at solute carrier family 16 (monocarboxylic acid SLC16A3 22q12.3-q13.2 -3.5 transporters), member 3 54152_at eukaryotic translation initiation factor 4E binding EIF4EBP1 8p12 -3.4 protein 1 43935_at ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin ARAP3 5q31.3 -3.2 homology domains-containing protein 3 33849_at pre-B-cell colony-enhancing factor PBEF 7q11.23 -3.2 46902_at N92294 -3.2 47023_at N25555 -3.1 Page 1 of 14 Supporting information for Sanoudou et al. -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Extensive Variation Between Inbred Mouse Strains Due to Endogenous L1 Retrotransposition
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition Keiko Akagi1,*, Jingfeng Li1,*, Robert M. Stephens3,*, Natalia Volfovsky3, and David E. Symer1,2,4 1) Basic Research Laboratory and 2) Laboratory of Biochemistry and Molecular Biology, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702; and 3) Advanced Biomedical Computing Center, Advanced Technology Program, SAIC-Frederick, Inc., Frederick, Maryland 21702 *these authors contributed equally to this work 4) to whom correspondence should be addressed at [email protected] Phone (301) 846 1560 Fax (301) 846 1638 Basic Research Laboratory, and Laboratory of Biochemistry and Molecular Biology, Building 560, Room 12-67, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702 Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Mouse variation from L1 retrotransposition Akagi et al. ABSTRACT. Numerous inbred mouse strains comprise models for human diseases and diversity, but the molecular differences between them are mostly unknown. Several mammalian genomes have been assembled, providing a framework for identifying structural variations. To identify variants between inbred mouse strains at a single nucleotide resolution, we aligned 26 million individual sequence traces from four laboratory mouse strains to the C57BL/6J reference genome. We discovered and analyzed over ten thousand intermediate-length genomic variants (from 100 nucleotides to 10 kilobases) distinguishing these strains from the C57BL/6J reference. Approximately 85% of such variants are due to recent mobilization of endogenous retrotransposons, predominantly L1 elements, greatly exceeding that reported in humans. -

Kinase Fusions Are Frequent in Spitz Tumours and Spitzoid Melanomas
ARTICLE Received 25 Sep 2013 | Accepted 15 Dec 2013 | Published 20 Jan 2014 DOI: 10.1038/ncomms4116 Kinase fusions are frequent in Spitz tumours and spitzoid melanomas Thomas Wiesner1,2,*, Jie He3,*, Roman Yelensky3,*, Rosaura Esteve-Puig4, Thomas Botton4,IweiYeh4, Doron Lipson3, Geoff Otto3, Kristina Brennan3, Rajmohan Murali5,6, Maria Garrido4, Vincent A. Miller3, Jeffrey S. Ross3, Michael F. Berger1, Alyssa Sparatta4, Gabriele Palmedo7, Lorenzo Cerroni2, Klaus J. Busam5, Heinz Kutzner7, Maureen T. Cronin3, Philip J. Stephens3 & Boris C. Bastian1,4,5 Spitzoid neoplasms are a group of melanocytic tumours with distinctive histopathological features. They include benign tumours (Spitz naevi), malignant tumours (spitzoid melano- mas) and tumours with borderline histopathological features and uncertain clinical outcome (atypical Spitz tumours). Their genetic underpinnings are poorly understood, and alterations in common melanoma-associated oncogenes are typically absent. Here we show that spitzoid neoplasms harbour kinase fusions of ROS1 (17%), NTRK1 (16%), ALK (10%), BRAF (5%) and RET (3%) in a mutually exclusive pattern. The chimeric proteins are constitutively active, stimulate oncogenic signalling pathways, are tumourigenic and are found in the entire biologic spectrum of spitzoid neoplasms, including 55% of Spitz naevi, 56% of atypical Spitz tumours and 39% of spitzoid melanomas. Kinase inhibitors suppress the oncogenic signalling of the fusion proteins in vitro. In summary, kinase fusions account for the majority of oncogenic aberrations in spitzoid neoplasms and may serve as therapeutic targets for metastatic spitzoid melanomas. 1 Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, 415 E 68th Street, New York, New York 10065, USA. 2 Department of Dermatology and Venereology, Medical University of Graz, Auenbruggerplatz 8, 8036 Graz, Austria.