Diversification Across Biomes in a Continental Lizard Radiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
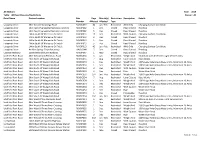
Year: 2016 Region: All Road Name Station Location Site Number
All Stations Year: 2016 Table: 4D Road Closures/Restrictions Region: All Road Name Station Location Site Days Month(s) Restriction Description Details Number Affected Affected Type Larapinta Drive 5Km West Of Areyonga Road RAVDC077 42 Jan - Feb Restricted 4Wd Only Changing Surface Conditions Larapinta Drive 1Km East Of Larapinta/Namatjira Junction RAVDP002 1 Dec Closed Road Closed Flooding Larapinta Drive 1Km East Of Larapinta/Namatjira Junction RAVDP002 5 Dec Closed Road Closed Flooding Larapinta Drive 14Km South Of Mereenie Oil Fields RAVDP013 15 Dec Restricted With Caution Changing Surface Conditions Larapinta Drive 14Km South Of Mereenie Oil Fields RAVDP013 2 Dec Closed Road Closed Flooding Larapinta Drive 14Km South Of Mereenie Oil Fields RAVDP013 1 Dec Closed Road Closed Flooding Larapinta Drive 14Km South Of Mereenie Oil Fields RAVDP013 5 Dec Closed Road Closed Flooding Larapinta Drive 14Km South Of Mereenie Oil Fields RAVDP013 42 Jan - Feb Restricted 4Wd Only Changing Surface Conditions Larapinta Drive At Alice Springs Town Boundary UAVDC044 5 Dec Closed Road Closed Flooding Lasseter Highway 500M West Of Stuart Highway RAVDP007 1 May Closed Road Closed Flooding Litchfield Park Road 2Km West Of Cox Peninsula Road RDVDC031 5 Jan Restricted Weight And Maximum Gvm 4.5 Tonne, Light Vehicles Only Litchfield Park Road 1Km North Of Wangi Falls Road RDVDC053 7 Aug Restricted Lane Closure Road Works Litchfield Park Road 1Km North Of Wangi Falls Road RDVDC053 1 Dec Restricted Weight And 100% Legal Axle Group Mass Limits, Maximum 13 Axles -

Driving Holidays in the Northern Territory the Northern Territory Is the Ultimate Drive Holiday Destination
Driving holidays in the Northern Territory The Northern Territory is the ultimate drive holiday destination A driving holiday is one of the best ways to see the Northern Territory. Whether you are a keen adventurer longing for open road or you just want to take your time and tick off some of those bucket list items – the NT has something for everyone. Top things to include on a drive holiday to the NT Discover rich Aboriginal cultural experiences Try tantalizing local produce Contents and bush tucker infused cuisine Swim in outback waterholes and explore incredible waterfalls Short Drives (2 - 5 days) Check out one of the many quirky NT events A Waterfall hopping around Litchfield National Park 6 Follow one of the unique B Kakadu National Park Explorer 8 art trails in the NT C Visit Katherine and Nitmiluk National Park 10 Immerse in the extensive military D Alice Springs Explorer 12 history of the NT E Uluru and Kings Canyon Highlights 14 F Uluru and Kings Canyon – Red Centre Way 16 Long Drives (6+ days) G Victoria River region – Savannah Way 20 H Kakadu and Katherine – Nature’s Way 22 I Katherine and Arnhem – Arnhem Way 24 J Alice Springs, Tennant Creek and Katherine regions – Binns Track 26 K Alice Springs to Darwin – Explorers Way 28 Parks and reserves facilities and activities 32 Festivals and Events 2020 36 2 Sealed road Garig Gunak Barlu Unsealed road National Park 4WD road (Permit required) Tiwi Islands ARAFURA SEA Melville Island Bathurst VAN DIEMEN Cobourg Island Peninsula GULF Maningrida BEAGLE GULF Djukbinj National Park Milingimbi -

Litchfield National Park
Litchfield National Park Litchfield National Park is an season only). Camping fees apply. Walkers, notify a reliable person of ancient landscape shaped by Generators are not permitted in your intended route and expected water. It features numerous Litchfield National Park return time. stunning waterfalls which A satellite phone or personal locator Accommodation, dining beacon is also recommended. cascade from the sandstone and camping - are also plateau of the Tabletop Range. available outside the Park at The Park covers approximately several commercial sites. Safety and Comfort 1500 sq km and contains Picnicking - shady spots • Swim only in designated areas. representative examples of most of available, see map. • Observe park safety signs. Fact Sheet the Top End’s natural habitats. • Carry and drink plenty of water. Cafe - located in the Wangi • Wear a shady hat, insect Intriguing magnetic termite Centre at Wangi Falls. mounds, historical sites and the repellent and sunscreen. weathered sandstone pillars of the Art Sales - Wangi Centre, • Wear suitable clothing and Lost City are a must for visitors. Wangi Falls. footwear. • Scrub Typhus is transmitted Whilst shady monsoon forest Swim - Florence Falls, walks provide retreats from the by microscopic bush mites Buley Rockhole, Wangi on grasses and bushes - avoid heat of the day. Falls, Walker Creek, Cascades, sitting on bare ground or grass. Aboriginal people have lived Tjaynera Falls and Surprise Creek • Carry a first aid kit. throughout the area for thousands Falls are designated swimming • Avoid strenuous activity during of years. It is important to areas. Note: some waterways can the heat of the day. the Koongurrukun, Mak Mak become unsafe after heavy rain • Note locations of Emergency Marranunggu, Werat and Warray and are closed for swimming - Call Devices. -

NT Seniors Card 2020-21 Business Discount Directory Information and Discounts for Territory Seniors
NT Seniors Card 2020-21 Business Discount Directory Information and discounts for Territory seniors www.ntseniorscard.org.au i 17% LIFETIME DISCOUNT* ON LIFE INSURANCE FOR NT SENIORS CARD MEMBERS Tourism NT/Shaana McNaught Why switch to NobleOak Life Insurance? Most awarded Australian Direct Life Insurer of 2019 Client satisfaction rating of 94.4%^ Comprehensive, fully-underwritten Life Insurance Lump sum payment if diagnosed with a terminal illness# Get an instant quote at: nobleoak.com.au/seniorscardnt Or call NobleOak for a quote: 1300 041 494 and mention ‘SENIORS CARD - NT’ to switch and save. NobleOak Life Limited ABN 85 087 648 708 AFSL No. 247302 issues the products. This information is of a general nature only and does not consider your individual objectives, financial situation or needs. Please consider the My Protection Plan Product Disclosure Statement (on website). Age limitations apply. People who seek to replace an existing Life Insurance policy should consider their circumstances including continuing the existing cover until the replacement policy is issued and cover confirmed. Online quotes are indicative only - actual premiums depend on factors such as health, age and pastimes. *Important information - savings information and discount. Considerable savings are possible - visit www.nobleoak.com.au/seniorscardnt/ for details of average savings on term life cover based on a premium comparison with life cover offered by a range of other Life Insurance companies undertaken in September 2019. Please note the premium comparison includes the 17% discount, which applies to usual term life cover premium rates. T&C apply (details on website) and the discount is on term life cover, available to Seniors Card Members (not in conjunction with a discount from any other program). -

Network Operating Guide Part A: Route Operating Protocols
Rail Safety Network Operating Guide Part A: Route Operating Protocols This document is uncontrolled unless s ta mp e d ‘ Controlled Do cu me n t ’ in red ink. This document is uncontrolled when copied or printed from an electronic version. Document number RS- NOG -032 PART A Re vis io n A Authorised by Scott MacGregor , General Manager Rail Safety Date of Issue 1 Au g u st 2016 THIS DOCUMENT REPLACES FL-PRO-06-005 PART A WHICH IS NOW OBSOLETE AND HAS BEEN REMOVED FROM THE GWA SAFETY MANAGEMENT SYSTEM This document is issued by Genesee and Wyoming Australia Pty Ltd The master copy of this manual is maintained electronically on the GWA Intranet site. Hard copies will NOT be centrally produced or distributed. Users who produce locally controlled hard copies of this manual should regularly check the issue status of the master on GWA Intranet site to ensure they are using the latest versions of these instructions, forms and procedures. COPYRIGHT. Subject to the Copyright Act, no SECTION of this manual may be reproduced by any process without the prior written permission from GWA's Director of Risk and Compliance. Function: Rail Safety Version No: 003 Document No: RS-NOG-032 Part A Issue Date: 01/08/2016 Document Uncontrolled When Copied or Printed RS-NOG-032 GWA Network Operating Guide Northgate BP to Berrimah Part A: Route Operating Protocols Amendments Page Issue Date of Amendment Details Number Number Issue All 001 26.06.2016 New document. Issued to replace (for 01.08.2016 FreightLink document FL-PRO-06-005 Part release) B which is now obsolete. -

Northern Territory Government Gazette No. G12, 23 March 2011
NORTHERN TERRITORY OF AUSTRALIA Government Gazette ISSN 0157 8324 No. G12 DARWIN 23 March 2011 GENERAL INFORMATION SUBSCRIPTIONS are payable in advance and are accepted for a maximum period of one calendar year. All subscriptions General issues of the Gazette contain notices under the are on a frm basis and refunds for cancellations will not be following headings: Proclamations; Legislative (Acts of Parliament assented to, Statutory Rules, By laws, given. Rates include surface postage in Australia and overseas. Regulations); Government departments administering Other carriage rates are available on application. particular legislation or functions; Notices under the Companies (Northern Territory) Code; Planning Act; Crown AVAILABILITY: The Gazette may be purchased by mail Lands Act; Private Notices; Tenders Invited; Contracts from: Awarded. Copies of each week’s General Gazette are available Retail Sales for a cost of $1.10 each (plus postage) and are published on a Wednesday. Copies of each week’s Special Gazettes are Government Printing Offce available separately for a cost of $1.10 each (plus postage). GPO Box 1447 Special Gazettes are supplied with General Gazettes on a Darwin NT 0801 Wednesday and they are sold at $1.10 per set (plus postage). Telephone: 08 8999 4031 Annual subscription rates apply from 1 July 2001. All current or purchased from paid subscriptions will not be effected and will continue until their expiry dates. Retail Sales Government Printing Offce NOTICES FOR PUBLICATION and related correspondence 203 Stuart Highway, Parap should be addressed to: Telephone: 08 8999 4031 Gazette Offce GPO Box 1447 Northern Territory Acts, Regulations and other Northern Darwin NT 0801 Territory Government legislation are only obtained from the Telephone: 08 8999 4005 Government Publications Offce, Darwin. -

Litchfield National Park
Northern Territory TOP END HOLIDAY GUIDE 2018 Edition DISCOVER TOP END DAY TOURS Kakadu National Park Explorer Litchfield National Park Waterfalls • Explore Kakadu National Park FULL DAY | D4 • Explore Litchfield National Park FULL DAY | D5 • Experience Warradjan Cultural Centre • Swim at Florence & Wangi Falls • Cruise the Yellow Water Billabong * • Visit Howard Springs Nature Park ADULT ADULT • Visit spectacular Nourlangie $265 • See spectacular Tolmer Falls $185 * • View ancient Aboriginal rock art $133 CHILD • Stop for a nice, relaxing lunch $93 CHILD Katherine Gorge Cruise & Edith Falls Culture, Wildlife & Wetlands • Explore Nitmiluk National Park FULL DAY | D11 • Cross the Adelaide River Floodplains FULL DAY | D6 • Cruise spectacular Katherine Gorge • Enjoy a lunch cruise on the Mary River Wetlands & Corroboree Billabong • Visit the Adelaide River War Cemetery * ADULT ADULT • Take a swim at Edith Falls $269 • Meet the local Indigenous people $209 * for insight on bush skills & tucker • See Aboriginal rock art $135 CHILD • Spot native flora & fauna $105 CHILD For more information please visit, Shop 6, 52 Mitchell Street, Darwin or call 1300 228 546 aatkings.com *Conditions: These prices do not include the entry fee to the Kakadu National Park. 12343a WELCOME TO THE TOP END Aboriginal people are the original custodians of the Top End and have a unique relationship with the land. Their art and Dreamtime stories weave a connection between spirit and country and provide modern travellers with a deeper understanding and insight to the mysteries of land and waters. The Northern Territory occupies Darwin is the Northern Territory’s hire with unlimited KM. about one sixth of Australia’s total multicultural capital, famed for its Further detailed information and land mass; its boundaries encompass markets and festivals, Asian cuisine advice is available by contacting a variety of contrasting landscapes and beautiful natural harbour. -

Coomalie Planning Concepts and Land Use Objectives 2000 Supports the Northern Territory Government’S Vision for Coomalie for the Next 20 to 25 Years
Department of Lands, Planning and Environment Foreword Part 1 - Planning Concepts of Coomalie Planning Concepts and Land Use Objectives 2000 supports the Northern Territory Government’s vision for Coomalie for the next 20 to 25 years. The Planning Concepts provide the direction and stimulus for public and private developers and the consent authority. This should facilitate the development of Coomalie in a manner which preserves its rural nature while recognising opportunities for continued development at a sustainable level. Part 1 - Planning Concepts provides the background information to Part 2 - Land Use Objectives. The Batchelor Division of the Northern Territory Planning Authority has been directed, and it is intended that any future Coomalie consent authority will be directed, to take the Planning Concepts into account along with those matters listed under the Planning Act 1993, when considering a development application. Part 2 - Land Use Objectives of Coomalie Planning Concepts and Land Use Objectives 2000 contains the land use objectives which have formal status by virtue of declaration under section 8(1) of the Act. Part 2 - Land Use Objectives establishes the framework for planning control within the Coomalie Sub- Region. To achieve this, land use objectives are provided under Key Objectives for the entire Sub-Region and under Land Use Objectives for specific land uses. The general discussion of issues assists in interpreting and implementing the objectives. The Coomalie Planning Concepts and Land Use Objectives 2000 aim to provide a co-ordinated and strategic approach to land use planning in Coomalie. In due course, the NT Planning Scheme Coomalie will apply to the entire Coomalie Sub-Region and parts of this Scheme will translate the land use objectives into planning practice. -

Northern Territory's Lithium Valley
NORTHERN TERRITORY’S LITHIUM VALLEY FINNISS LITHIUM PROJECT MINING THE TERRITORY CONFERENCE Darwin September 2018 ASX:CXO DISCLAIMER The information in this report that relates to Exploration Results and Mineral Resources is based on information compiled by Stephen Biggins (BSc(Hons)Geol, MBA) as Managing Director of Core Exploration Ltd who is a member of the Australasian Institute of Mining and Metallurgy and is bound by and follows the Institute’s codes and recommended practices. He has sufficient experience which is relevant to the styles of mineralisation and types of deposits under consideration and to the activities being undertaken to qualify as a Competent Person as defined in the 2012 Edition of the “Australasian Code for Reporting of Exploration Results, Mineral Resources and Ore Reserves”. Mr. Biggins consents to the inclusion in the report of the matters based on his information in the form and context in which it appears. This document has been prepared by Core Exploration Limited (“Core”, “Company”) and provided as a basic overview of the tenements held or controlled by the Company. This presentation does not purport to be all-inclusive or to contain all the information that you or any other party may require to evaluate the prospects of the Company. None of the Company, any of its related bodies corporate or any of their representatives assume any responsibility for, or makes any representation or warranty, express or implied, with respect to the accuracy, reliability or completeness of the information contained in this document and none of those parties have or assume any obligation to provide any additional information or to update this document. -

2013 Power and Water Corporation Darwin Region Water Supply Strategy
POWER AND WATER CORPORATION DARWIN REGION WATER SUPPLY STRATEGY 2013 Darwin Region Water Supply Strategy #1080991/04-2015 power and water corporation Darwin Region Water Supply Strategy i Contents 1 EXECUTIVE SUMMARY................................................................................................................................1 3.7 Emergency Supply............................................................................................................................... 18 1.1 Introduction............................................................................................................................................................1 3.8 Water Demand Trends............................................................................................................... 19 . 1.2 Water Supply Security and Sustainability............................................1 3.9 Water Supply Pricing..................................................................................................................... 20 1.2.1. Risk.–.Sustainability.of.Supply.................................................................1 4 WATER DEMAND FORECASTING.................................................................................21 1.2.2. Mitigation.–.Sustainability.of.Supply.......................................1 4.1 Introduction...................................................................................................................................................... 21 1.2.3. Risk.–.Security.of.Supply........................................................................................1 -

Marsupial' Freshwater
Zoosyst. Evol. 85 (2) 2009, 199–275 / DOI 10.1002/zoos.200900004 Diversity and disparity ‘down under’: Systematics, biogeography and reproductive modes of the ‘marsupial’ freshwater Thiaridae (Caenogastropoda, Cerithioidea) in Australia Matthias Glaubrecht*,1, Nora Brinkmann2 and Judith Pppe1 1 Museum fr Naturkunde Berlin, Department of Malacozoology, Invalidenstraße 43, 10115 Berlin, Germany 2 University of Copenhagen, Institute of Biology, Research Group for Comparative Zoology, Universitetsparken 15, 2100 Copenhagen, Denmark Abstract Received 11 May 2009 We systematically revise here the Australian taxa of the Thiaridae, a group of freshwater Accepted 15 June 2009 Cerithioidea with pantropical distribution and “marsupial” (i.e. viviparous) reproductive Published 24 September 2009 modes. On this long isolated continent, the naming of several monotypic genera and a plethora of species have clouded both the phylogenetical and biogeographical relation- ships with other thiarids, in particular in Southeast Asia, thus hampering insight into the evolution of Australian taxa and their natural history. Based on own collections during five expeditions to various regions in Australia between 2002 and 2007, the study of rele- vant type material and the comparison with (mostly shell) material from major Australian museum collections, we describe and document here the morphology (of adults and juve- niles) and radulae of all relevant thiarid taxa, discussing the taxonomical implications and nomenclatural consequences. Presenting comprehensive compilations of the occurrences for all Australian thiarid species, we document their geographical distribution (based on over 900 records) with references ranging from continent-wide to drainage-based pat- terns. We morphologically identify a total of eleven distinct species (also corroborated as distinct clades by molecular genetic data, to be reported elsewhere), of which six species are endemic to Australia, viz. -
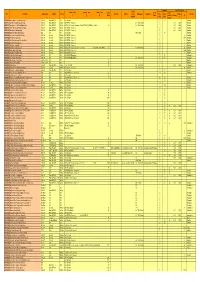
NT Appendix 2B.1
Sampling HydroTel Polling Shelter Sensor Type Sensor Type Sensor Type Sensor Solar Hours Site Site Name Data Logger Modem Comms Platform Shelter Serial Rain Gauge Regulator Time Stage Afternoo Site Type Range Panel with no Morning IP 1 2 3 Number (min) (mm) n change G0010005 Ranken River at Soudan Homestead iRIS 350 Beam RST600 N/A Shaft Encoder 10 10 Reactive G0050115 Hugh River at South Road Crossing iRIS 350 Beam RST600 DialUp HS AD375A ( Absolute) HS TB 2 (0.5mm) 10 6:30 14:30 Reactive G0050116 Finke River at South Road Bridge X-ing iRIS 350 Beam RST600 DialUp HS WL3100 Pressure Transducer Druck PTX 1400 (250 Bar) HS 23 SL 10m HS TB 3 (0.5mm) 10 6:30 14:30 Reactive G0050117 Palmer River at South Road Crossing iRIS 350 Beam RST600 DialUp HS AD375A ( Absolute) 15 6:30 14:30 Reactive G0050140 Finke River at Railway Bridge iRIS 350 Beam RST600 DialUp HS AD375A ( Absolute) 15 6:30 14:30 Reactive G0060005 Trephina Creek at Trephina Gorge SDS N/A N/A Shaft Encoder TB1 (0.5mm 10 10 Reactive G0060008 Roe Creek at South Road Crossing iRIS 320 (internal) IP Mode HS AD375A ( Absolute) 10 1h Reactive G0060009 Todd River at Anzac Oval iRIS 320 (internal) IP Mode HS AD375A ( Absolute) 10 30min Reactive G0060017 Emily Creek Upstream Undoolya Road iRIS 320 (internal) IP Mode HS AD375A ( Absolute) 10 1h Reactive G0060040 Todd River at Amoonguna iRIS 320 (internal) IP Mode HS AD375A ( Absolute) 10 1h Reactive G0060041 Todd River Rocky Hill iRIS 320 (internal) IP Mode HS AD375A ( Absolute) 10 1h Reactive G0060046 Todd River at Wigley Gorge iRIS 320 (internal)