Correcting Improper Chromosome–Spindle Attachments During Cell Division
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chromosome Missegregation in Human Cells Arises Through Specific
Chromosome missegregation in human cells arises through specific types of kinetochore–microtubule attachment errors Sarah L. Thompson and Duane A. Compton1 Department of Biochemistry, Dartmouth Medical School, Hanover, NH 03755; and Cancer Mechanisms Program, Norris Cotton Cancer Center, Lebanon, NH 03766 Edited by John W. Sedat, University of California, San Francisco School of Medicine, San Francisco, CA, and approved September 15, 2011 (received for review June 17, 2011) Most solid tumors are aneuploid, and many missegregate chro- cur, particularly in early phases of mitosis, as a consequence mosomes at high rates in a phenomenon called chromosomal of the stochastic interactions between microtubules and kinet- instability (CIN). CIN reflects the erosion of mitotic fidelity, and it ochores (11). A prominent error is when a single kinetochore correlates with poor patient prognosis and drug resistance. The binds microtubules oriented toward both spindle poles. This most common mechanism causing CIN is the persistence of improper error is called merotely (12, 13). The persistence of merotely kinetochore–microtubule attachments called merotely. Chromo- undermines chromosome segregation because merotelic kinet- somes with merotelic kinetochores often manifest as lagging chro- ochores experience poleward force toward both spindle poles. As mosomes in anaphase, suggesting that lagging chromosomes fail a consequence, merotely often results in the appearance of lag- to segregate properly. However, it remains unknown whether the ging chromosomes in anaphase, and tumor cells with CIN have lagging chromosomes observed in anaphase segregate to the cor- elevated rates of lagging chromosomes and merotelic attach- rect or incorrect daughter cell. To address this question, we tracked ments (9). Moreover, it was shown that increasing the correction the segregation of a single human chromosome during cell division rate of merotely by stimulating the dynamics of k-MT attachment by using LacI-GFP to target an integrated LacO array. -

Prevalence of Linear Configuration Among Chain Trivalents at Metaphase I in Pollen Mother Cells of Petunia Axillaris (LAM.) B
_??_1991 by Cytologia, Tokyo Cytologia 56: 367 -371 , 1991 Prevalence of Linear Configuration among Chain Trivalents at Metaphase I in Pollen Mother Cells of Petunia axillaris (LAM.) B. S. P. P. China Pullaiah, P. S. R. L. Narasinga Rao and V. Padmaja Department of Botany, Andhra University, Waltair, India Accepted February 28, 1991 At somatic metaphase the kinetochore of each sister chromatid interacts with the spindle in such a way that the two subsequently disjoin one to each pole. At metaphase I of meiosis in disomics, a pair of chromatids is one half of a bivalent. Here, in their involvement with the spindle, the two kinetochores of a chromatid pair are not poised disjunctionally, and the centromere is thus said to be syntelic. For a 'double kinetochore' of each member of a meiotic bivalent, 'syntelic' aspect is thus a derived state. Failure of the 'double kinetochore' to attain to 'syntelic' state is rare for the members of a bivalent. On the other hand, the 'kinetochore pair' of a univalent at meiosis I may assume an amphitelic state at metaphase I even if the initial development was towards a syntelic state (Bauer et al. 1961, Sybenga 1975). When we consider chain trivalents, which indeed are the simplest of multivalents, syntelic state for all three kinetochore pairs is quite prevalent and is often seen in the alternate configuration where the mid-member is poised disjunctionally from both its neighbours. Much rarer is the variant, where the three syntelic centromeres disjoin in a 2-1 manner as in the case of ad jacent orientation. -

The Mitotic Checkpoint Kinase NEK2A Regulates Kinetochore Microtubule Attachment Stability
Oncogene (2008) 27, 4107–4114 & 2008 Nature Publishing Group All rights reserved 0950-9232/08 $30.00 www.nature.com/onc ORIGINAL ARTICLE The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability JDu1,6, X Cai1,2,6, J Yao1, X Ding2,3,QWu1,2, S Pei1, K Jiang1,2, Y Zhang1, W Wang3, Y Shi1, Y Lai1, J Shen1, M Teng1, H Huang4, Q Fei5, ES Reddy2, J Zhu5, C Jin1 and X Yao1,2 1Hefei National Laboratory for Physical Sciences at Micro-scale, University of Science and Technology of China, Hefei, China; 2Cancer Biology Program, Morehouse School of Medicine, Atlanta, GA, USA; 3Department of Medicine, Beijing University of Chinese Medicine, 4Department of Hematology, the 1st Affiliated Hospital, Zhejiang University, Hongzhou, China and 5Cancer Epigenetics Program, Shanghai Cancer Institute, Shanghai Jiaotong University, Shanghai, China Loss or gain of whole chromosome, the form of Introduction chromosome instability commonly associated with cancers is thought to arise from aberrant chromosome segregation Chromosomal instability (CIN) has been recognized as a during cell division. Chromosome segregation in mitosis is hallmark of human cancer and is caused by continuous orchestrated by the interaction of kinetochores with chromosome missegregation during cell division. Proper spindle microtubules. Our studies showthat NEK2A is a chromosome segregation requires a faithful physical link kinetochore-associated protein kinase essential for faith- between spindle microtubules and centromeric DNA via ful chromosome segregation. However, it was unclear how a protein supercomplex called kinetochore (Cleveland NEK2A ensures accurate chromosome segregation in et al., 2003). In addition to providing a physical link mitosis. Here we show that NEK2A-mediated Hec1 between chromosomes and spindle microtubules, the (highly expressed in cancer) phosphorylation is essential kinetochore has an active function in orchestrating for faithful kinetochore microtubule attachments in chromosome movements through microtubule motors mitosis. -

A Stochastic Model of Kinetochore–Microtubule Attachment Accurately
A stochastic model of kinetochore–microtubule attachment accurately describes fission yeast chromosome segregation Guillaume Gay, Thibault Courthéoux, Céline Reyes, Sylvie Tournier, Yannick Gachet To cite this version: Guillaume Gay, Thibault Courthéoux, Céline Reyes, Sylvie Tournier, Yannick Gachet. A stochas- tic model of kinetochore–microtubule attachment accurately describes fission yeast chromosome segregation. Journal of Cell Biology, Rockefeller University Press, 2012, 196 (6), pp.757-774. 10.1083/jcb.201107124. hal-02380748 HAL Id: hal-02380748 https://hal.archives-ouvertes.fr/hal-02380748 Submitted on 26 Nov 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Published March 12, 2012 JCB: Article A stochastic model of kinetochore–microtubule attachment accurately describes fission yeast chromosome segregation Guillaume Gay,1,2 Thibault Courtheoux,1,2 Céline Reyes,1,2 Sylvie Tournier,1,2 and Yannick Gachet1,2 1Laboratoire de biologie cellulaire et moléculaire du contrôle de la proliferation, Université de Toulouse, F-31062 Toulouse, France 2Unité Mixte de Recherche 5088, Centre National de la Recherche Scientifique, F-31062 Toulouse, France n fission yeast, erroneous attachments of spindle micro segregation seen in fission yeast. Prevention of attachment tubules to kinetochores are frequent in early mitosis. -

Identification of a Global Aneuploidy-Associated Transcriptional and Phenotypic Signature in Budding Yeast
IDENTIFICATION OF A GLOBAL ANEUPLOIDY-ASSOCIATED TRANSCRIPTIONAL AND PHENOTYPIC SIGNATURE IN BUDDING YEAST by Anjali Nelliat A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Master of Science in Engineering Baltimore, Maryland May, 2017 Abstract Aneuploidy, an unbalanced genome state with either gain or loss of chromosomes, is known to have dramatic effects on cellular physiology, ranging from detrimental effects including genetic disorders such as Down Syndrome to promoting adaptability under stress, such as drug resistance. This paradoxical nature of aneuploidy impedes the development of therapeutic strategies since a vast majority of tumors exhibit complex aneuploid karyotypes. Although the aneuploid cancer genome has been extensively studied, common transcriptional and phenotypic consequences of aneuploidy still remain elusive. To comprehensively study aneuploidy-specific features, we first generated karyotypically heterogeneous aneuploid populations in budding yeast Saccharomyces cerevisiae, based on our general statistical model. We then performed whole transcriptome sequencing on these heterogeneous aneuploid populations, to uncover a karyotype- independent, global aneuploidy-associated transcriptional response. The aneuploid transcriptome was similar to gene expression profiles of yeast cells subject to hypo-osmotic and cell-wall stresses. Furthermore, we observed that aneuploid cells indeed exhibited phenotypes characteristic of these stresses, including cell swelling, increased -

Modeling the Temporal Evolution of the Spindle Assembly Checkpoint and Role of Aurora B Kinase
Modeling the temporal evolution of the spindle assembly checkpoint and role of Aurora B kinase Hitesh B. Mistrya, David E. MacCallumb, Robert C. Jacksonb, Mark A. J. Chaplaina, and Fordyce A. Davidsona,1 aDivision of Mathematics, University of Dundee, Dundee DD1 4HN, Scotland; and bCyclacel Ltd., Dundee Technopole, Dundee DD1 5JJ, Scotlandb; Communicated by Avner Friedman, Ohio State University, Columbus, OH, November 3, 2008 (received for review March 11, 2008) Faithful separation of chromosomes prior to cell division at mito- In budding yeast Ipl1 can promote correct spindle assembly by sis is a highly regulated process. One family of serine/threonine destabilizing syntelic attachments (22, 23). This has also been kinases that plays a central role in regulation is the Aurora family. demonstrated in mammalian cells for syntelic attachments (14, 24) Aurora B plays a role in the spindle assembly checkpoint, in part, by and merotelic attachments (25). Aurora B promotes turnover of destabilizing the localization of BubR1 and Mad2 at centrosomes microtubules at the kinetochores (21) possibly by regulating Hec and responds to changes in tension caused by aberrant microtubule 1 (26). Aurora B is also required for cytokinesis where it phospho- kinetochore attachments. Aurora B is overexpressed in a subset of rylates and regulates several substrates (7, 8). Characterization of cancers and is required for mitosis, making it an attractive anti- Aurora B inhibitors has suggested that Aurora B plays a role in cancer target. Here, we use mathematical modeling to extend a the spindle assembly checkpoint, in part, by destabilizing the local- current model of the spindle assembly checkpoint to incorporate ization of BuBR1, Mad2, and Cenp-E at centrosomes (14, 27) all signaling kinetochores within a cell rather than just one and the and responds to changes in tension to promote biorientation role of Aurora B within the resulting model. -

Enhancing Togetherness: Kinetochores and Cohesion
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE Enhancing togetherness: kinetochores and cohesion Jennifer L. Gerton1 Stowers Institute for Medical Research and University of Kansas Medical Center, Kansas City, Missouri 64110, USA All organisms must correctly copy and equally distribute two different cases of this: syntelic and monotelic. In their genetic information each and every time a new cell syntelic attachment, both sister kinetochores are at- is created. In eukaryotic cells, this action involves many tached to microtubules from the same pole (Fig. 1B). In different coordinated processes, including DNA replica- monotelic attachment, one of the kinetochores may be tion, kinetochore and spindle formation, and chromo- attached to microtubules from one pole and the other some cohesion. If errors occur in chromosome segrega- kinetochore is unattached (Fig. 1C). In a merotelic at- tion, a chromosome may be lost or gained, resulting in tachment, one kinetochore may be attached to microtu- an aneuploid cell. Aneuploidy is associated with cancer bules from both poles. This occurs in organisms that in humans, as well as a variety of developmental disor- have multiple microtubules attached to a single kineto- ders including Downs syndrome. Because all organisms chore. Defects in kinetochore attachment may lead to require the faithful transmission of their genetic infor- the activation of the spindle checkpoint. mation, many of these processes and proteins are evolu- All eukaryotic cells studied to date have a mitotic tionarily conserved. Thus, gaining a basic understanding spindle checkpoint. This refers to the fact that defects in of the basic mechanisms that contribute to the accurate the mitotic spindle trigger a cascade of events that lead segregation of chromosomes is essential for understand- to cell cycle delay or arrest. -
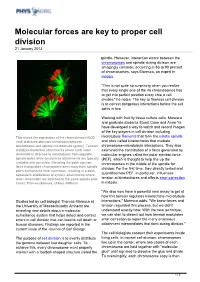
Molecular Forces Are Key to Proper Cell Division 21 January 2013
Molecular forces are key to proper cell division 21 January 2013 spindle. However, interaction errors between the chromosomes and spindle during division are amazingly common, occurring in 86 to 90 percent of chromosomes, says Maresca, an expert in mitosis. "This is not quite so surprising when you realize that every single one of the 46 chromosomes has to get into perfect position every time a cell divides," he notes. The key to flawless cell division is to correct dangerous interactions before the cell splits in two. Working with fruit fly tissue culture cells, Maresca and graduate students Stuart Cane and Anna Ye have developed a way to watch and record images of the key players in cell division including This shows the expression of the chromokinesin NOD microtubule filaments that form the mitotic spindle (red) stabilizes aberrant interactions between and sites called kinetochores that mediate kinetochores and spindle microtubules (green). Tension chromosome-microtubule interactions. They also stabilizes bioriented attachments where each sister examined the contribution of a force generated by chromatid is attached to microtubules from opposite molecular engines called the polar ejection force spindle poles while tensionless attachments are typically (PEF), which is thought to help line up the unstable and corrected. Elevating the polar ejection chromosomes in the middle of the spindle for force that pushes chromosome arms away from spindle division. For the first time, they directly tested and poles overwhelms error correction, resulting in a dose- dependent stabilization of syntelic attachments where quantified how PEF, in particular, influences sister chromatids are attached to the same spindle pole. -

Spindle Dynamics Model Explains Chromosome Loss Rates in Yeast Polyploid Cells
HYPOTHESIS AND THEORY published: 06 August 2018 doi: 10.3389/fgene.2018.00296 Spindle Dynamics Model Explains Chromosome Loss Rates in Yeast Polyploid Cells Ivan Jelenic´ 1, Anna Selmecki 2, Liedewij Laan 3* and Nenad Pavin 1* 1 Department of Physics, Faculty of Science, University of Zagreb, Zagreb, Croatia, 2 Department of Medical Microbiology and Immunology, Creighton University Medical School, Omaha, NE, United States, 3 Department of Bionanoscience, Faculty of Applied Sciences, Kavli Institute of NanoScience, Delft University of Technology, Delft, Netherlands Faithful chromosome segregation, driven by the mitotic spindle, is essential for organismal survival. Neopolyploid cells from diverse species exhibit a significant increase in mitotic errors relative to their diploid progenitors, resulting in chromosome nondisjunction. In the model system Saccharomyces cerevisiae, the rate of chromosome loss in haploid and diploid cells is measured to be one thousand times lower than the rate Edited by: of loss in isogenic tetraploid cells. Currently it is unknown what constrains the number Richard John Abbott, of chromosomes that can be segregated with high fidelity in an organism. Here we University of St Andrews, developed a simple mathematical model to study how different rates of chromosome United Kingdom loss in cells with different ploidy can arise from changes in (1) spindle dynamics and Reviewed by: Pirita Paajanen, (2) a maximum duration of mitotic arrest, after which cells enter anaphase. We apply John Innes Centre (JIC), this model to S. cerevisiae to show that this model can explain the observed rates of United Kingdom Yongshuai Sun, chromosome loss in S. cerevisiae cells of different ploidy. -

The Consequences of Tetraploidy and Aneuploidy
Commentary 3859 The consequences of tetraploidy and aneuploidy Zuzana Storchova* and Christian Kuffer Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany *Author for correspondence (e-mail: [email protected]) Accepted 16 October 2008 Journal of Cell Science 121, 3859-3866 Published by The Company of Biologists 2008 doi:10.1242/jcs.039537 Summary Polyploidy, an increased number of chromosome sets, is a stable aneuploidy are commonly observed in cancers. Recently, surprisingly common phenomenon in nature, particularly in it has been proposed that an increased number of chromosome plants and fungi. In humans, polyploidy often occurs in specific sets can promote cell transformation and give rise to an tissues as part of terminal differentiation. Changes in ploidy aneuploid tumor. Here, we review how tetraploidy can occur can also result from pathophysiological events that are caused and describe the cellular responses to increased ploidy. by viral-induced cell fusion or erroneous cell division. Furthermore, we discuss how the specific physiological changes Tetraploidization can initiate chromosomal instability (CIN), that are triggered by polyploidization might be used as novel probably owing to supernumerary centrosomes and the doubled targets for cancer therapy. chromosome mass. CIN, in turn, might persist or soon give way to a stably propagating but aneuploid karyotype. Both CIN and Key words: Aneuploidy, Chromosomal instability, Tetraploidy Introduction aneuploidy (Chi and Jeang, 2007; Kops et al., 2005), but an Most malignant tumors have been found to have an abnormal alternative and more radical mechanism might exist in which karyotype with multiple structural and numerical aberrations of inherently instable tetraploid cells can evolve into tumorigenic chromosomes – so-called ‘aneuploidy’. -

The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases
cells Review The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases 1, , 2 1, , Francesca Degrassi * y , Michela Damizia and Patrizia Lavia * y 1 IBPM Institute of Molecular Biology and Pathology, CNR Consiglio Nazionale delle Ricerche, c/o Department of Biology and Biotechnology “Charles Darwin”, Sapienza University of Rome, 00185 Roma, Italy 2 Department of Biology and Biotechnology “Charles Darwin”, Sapienza University of Rome, 00185 Roma, Italy; [email protected] * Correspondence: [email protected] (F.D.); [email protected] (P.L.); Tel.: +39-06-49917517 (F.D.); +39-06-49917536 (P.L.) These authors contributed equally to this work. y Received: 20 November 2019; Accepted: 21 December 2019; Published: 24 December 2019 Abstract: Regulators of mitotic division, when dysfunctional or expressed in a deregulated manner (over- or underexpressed) in somatic cells, cause chromosome instability, which is a predisposing condition to cancer that is associated with unrestricted proliferation. Genes encoding mitotic regulators are growingly implicated in neurodevelopmental diseases. Here, we briefly summarize existing knowledge on how microcephaly-related mitotic genes operate in the control of chromosome segregation during mitosis in somatic cells, with a special focus on the role of kinetochore factors. Then, we review evidence implicating mitotic apparatus- and kinetochore-resident factors in the origin of congenital microcephaly. We discuss data emerging from these works, which suggest a critical role of correct mitotic division in controlling neuronal cell proliferation and shaping the architecture of the central nervous system. Keywords: microcephaly; mitotic apparatus; kinetochore; chromosome segregation; neural progenitors 1. Introduction Regulators of the mitotic apparatus play key roles in orchestrating chromosome segregation. -

Aurora Kinases: Shining Lights on the Therapeutic Horizon?
Oncogene (2005) 24, 5005–5015 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc REVIEW Aurora kinases: shining lights on the therapeutic horizon? Paul D Andrews*,1 1Division of Gene Regulation and Expression, Wellcome Trust Biocentre, University of Dundee, Dundee, DD1 5EH, UK The Aurora kinases have been implicated in tumorigenesis to ensure accurate chromosome segregation and timely and are important regulators of diverse cell cycle events, cytokinesis. The approximate execution points of ranging from the entry into mitosis,centrosome function, Aurora A and B are shown in Figure 2. Aurora C’s mitotic spindle formation,chromosome biorientation and regulation and localization have remained obscure until segregation,and cytokinesis. The recent identification of recently (Li et al., 2004b; Sasai et al., 2004) and its real novel binding partners and key downstream effectors, function is unknown. together with new small-molecule inhibitors that display With over protein 500 kinases in the human genome efficacy against tumours,heralds an upsurge of interest in grouped into at least 20 structurally related families, the these critical kinases. This review details new develop- specificity of kinase inhibitors is a critical concern when ments in the field and analyses the potential of Aurora administrated in therapy. However, the recognition that kinases as anticancer targets. the selective inhibition of individual protein kinases is Oncogene (2005) 24, 5005–5015. doi:10.1038/sj.onc.1208752 not only possible, but has therapeutic efficacy (as exemplified by the success of the Bcl-Abl kinase Keywords: Aurora kinase; microtubule; spindle inhibitor, Gleevec, for the treatment of chronic myeloid checkpoint; centrosome; kinetochore; therapy leukemia) has led to a huge interest in the development of kinase inhibitors for cancer therapy.