Structure Based Drug Designing for Diabetes Mellitus K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolic-Hydroxy and Carboxy Functionalization of Alkyl Moieties in Drug Molecules: Prediction of Structure Influence and Pharmacologic Activity
molecules Review Metabolic-Hydroxy and Carboxy Functionalization of Alkyl Moieties in Drug Molecules: Prediction of Structure Influence and Pharmacologic Activity Babiker M. El-Haj 1,* and Samrein B.M. Ahmed 2 1 Department of Pharmaceutical Sciences, College of Pharmacy and Health Sciences, University of Science and Technology of Fujairah, Fufairah 00971, UAE 2 College of Medicine, Sharjah Institute for Medical Research, University of Sharjah, Sharjah 00971, UAE; [email protected] * Correspondence: [email protected] Received: 6 February 2020; Accepted: 7 April 2020; Published: 22 April 2020 Abstract: Alkyl moieties—open chain or cyclic, linear, or branched—are common in drug molecules. The hydrophobicity of alkyl moieties in drug molecules is modified by metabolic hydroxy functionalization via free-radical intermediates to give primary, secondary, or tertiary alcohols depending on the class of the substrate carbon. The hydroxymethyl groups resulting from the functionalization of methyl groups are mostly oxidized further to carboxyl groups to give carboxy metabolites. As observed from the surveyed cases in this review, hydroxy functionalization leads to loss, attenuation, or retention of pharmacologic activity with respect to the parent drug. On the other hand, carboxy functionalization leads to a loss of activity with the exception of only a few cases in which activity is retained. The exceptions are those groups in which the carboxy functionalization occurs at a position distant from a well-defined primary pharmacophore. Some hydroxy metabolites, which are equiactive with their parent drugs, have been developed into ester prodrugs while carboxy metabolites, which are equiactive to their parent drugs, have been developed into drugs as per se. -

Sulfonylureas
Therapeutic Class Overview Sulfonylureas INTRODUCTION In the United States (US), diabetes mellitus affects more than 30 million people and is the 7th leading cause of death (Centers for Disease Control and Prevention [CDC] 2018). Type 2 diabetes mellitus (T2DM) is the most common form of diabetes and is characterized by elevated fasting and postprandial glucose concentrations (American Diabetes Association [ADA] 2019[a]). It is a chronic illness that requires continuing medical care and ongoing patient self-management education and support to prevent acute complications and to reduce the risk of long-term complications (ADA 2019[b]). ○ Complications of T2DM include hypertension, heart disease, stroke, vision loss, nephropathy, and neuropathy (ADA 2019[a]). In addition to dietary and lifestyle management, T2DM can be treated with insulin, one or more oral medications, or a combination of both. Many patients with T2DM will require combination therapy (Garber et al 2019). Classes of oral medications for the management of blood glucose levels in patients with T2DM focus on increasing insulin secretion, increasing insulin responsiveness, or both, decreasing the rate of carbohydrate absorption, decreasing the rate of hepatic glucose production, decreasing the rate of glucagon secretion, and blocking glucose reabsorption by the kidney (Garber et al 2019). Pharmacologic options for T2DM include sulfonylureas (SFUs), biguanides, thiazolidinediones (TZDs), meglitinides, alpha-glucosidase inhibitors, dipeptidyl peptidase-4 (DPP-4) inhibitors, glucagon-like peptide-1 (GLP-1) analogs, amylinomimetics, sodium-glucose cotransporter 2 (SGLT2) inhibitors, combination products, and insulin (Garber et al 2019). SFUs are the oldest of the oral antidiabetic medications, and all agents are available generically. The SFUs can be divided into 2 categories: first-generation and second-generation. -

(Glyburide) Tablets USP 1.25, 2.5 and 5 Mg DESCRIPTION Diaßeta
Diaßeta® (glyburide) Tablets USP 1.25, 2.5 and 5 mg DESCRIPTION Diaßeta® (glyburide) is an oral blood-glucose-lowering drug of the sulfonylurea class. It is a white, crystalline compound, formulated as tablets of 1.25 mg, 2.5 mg, and 5 mg strengths for oral administration. Diaßeta tablets USP contain the active ingredient glyburide and the following inactive ingredients: dibasic calcium phosphate USP, magnesium stearate NF, microcrystalline cellulose NF, sodium alginate NF, talc USP. Diaßeta 1.25 mg tablets USP also contain D&C Yellow #10 Aluminum Lake and FD&C Red #40 Aluminum Lake. Diaßeta 2.5 mg tablets USP also contain FD&C Red #40 Aluminum Lake. Diaßeta 5 mg tablets USP also contain D&C Yellow #10 Aluminum Lake, and FD&C Blue #1. Chemically, Diaßeta is identified as 1-[[p-[2-(5-Chloro-o-anisamido)ethyl]phenyl]sulfonyl]-3-cyclohexylurea. The CAS Registry Number is 10238-21-8. The structural formula is: Cl CONHCH2CH2 SO2NHCONH OCH3 The molecular weight is 493.99. The aqueous solubility of Diaßeta increases with pH as a result of salt formation. CLINICAL PHARMACOLOGY Diaßeta appears to lower the blood glucose acutely by stimulating the release of insulin from the pancreas, an effect dependent upon functioning beta cells in the pancreatic islets. The mechanism by which Diaßeta lowers blood glucose during long-term administration has not been clearly established. With chronic administration in Type II diabetic patients, the blood glucose lowering effect persists despite a gradual decline in the insulin secretory response to the drug. Extrapancreatic effects may play a part in the mechanism of action of oral sulfonylurea hypoglycemic drugs. -

In Vitro Inhibition of Breast Cancer Spheroid-Induced Lymphendothelial
FULL PAPER British Journal of Cancer (2013) 108, 570–578 | doi: 10.1038/bjc.2012.580 Keywords: lymphendothelial intravasation; tumour spheroid; acetohexamide; nifedipin; isoxsuprine; proadifen In vitro inhibition of breast cancer spheroid-induced lymphendothelial defects resembling intravasation into the lymphatic vasculature by acetohexamide, isoxsuprine, nifedipin and proadifen N Kretschy1,8, M Teichmann1,8, S Kopf1, A G Atanasov2, P Saiko3, C Vonach1, K Viola1, B Giessrigl1, N Huttary1, I Raab1, S Krieger1,WJa¨ ger4, T Szekeres3, S M Nijman5, W Mikulits6, V M Dirsch2, H Dolznig7, M Grusch6 and G Krupitza*,1 1Institute of Clinical Pathology, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; 2Department of Pharmacognosy, University of Vienna, Althanstrasse 14, A-1090 Vienna, Austria; 3Clinical Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; 4Department for Clinical Pharmacy and Diagnostics, Faculty of Life Sciences, University of Vienna, Althanstrasse 14, A-1090 Vienna, Austria; 5Research Center for Molecular Medicine of the Austrian Academy of Sciences, Lazarettgasse 14, AKH BT 25.3, A-1090 Vienna, Austria; 6Department of Medicine I, Institute of Cancer Research, Medical University of Vienna, Borschkegasse 8a, A-1090 Vienna, Austria and 7Institute of Medical Genetics, Medical University of Vienna, Waehringer Strasse 10, A-1090 Vienna, Austria Background: As metastasis is the prime cause of death from malignancies, there is vibrant interest to discover options for the management of the different mechanistic steps of tumour spreading. Some approved pharmaceuticals exhibit activities against diseases they have not been developed for. In order to discover such activities that might attenuate lymph node metastasis, we investigated 225 drugs, which are approved by the US Food and Drug Administration. -

Oral Hypoglycemic Agents
Medication Instructions Oral Hypoglycemic Agents GENERIC BRAND Glimepiride Amaryl Glyburide DiaBeta, Micronase, Glynase Glipizide Glucotrol, GlucotrolXL Repaglinide Prandin Acetohexamide Dymelor Chlorpropamide Diabinese Tolazamide Tolinase Tolbutamide Orinase Description Oral hypoglycemic agents (OHAs) are a group of drugs used to help reduce the amount of sugar present in the blood. They are not insulin, but they stimulate the pancreas to produce insulin. OHAs are usually used in the treatment of adult onset diabetes (also known as Type 2 or non-insulin dependent diabetes mellitus). REMEMBER THIS MEDICINE WILL NOT CURE YOUR DIABETES, BUT DOES HELP TO CONTROL IT. The OHA, along with your meal and exercise plans should help you reach your target blood sugar. A stable blood glucose level helps to reduce the risk of heart, kidney, eye and blood vessel diseases. Check with your doctor to find out your target blood sugar range. Dosage and Administration Take this medicine 30 minutes before a meal and at about the same time every day. If it causes stomach upset, then take it with the meal. Try not to skip meals when taking these medicines. Your doctor will decide on the amount of medicine you will need and how often you should take it. If you miss a dose, take it as soon as possible. If it is almost time for your next dose, do not take the missed dose. Do not double the next dose. Instead, go back to your regular dosing schedule. Side Effects The most common side effect of OHAs is low blood sugar (hypoglycemia). Each person’s reaction to low blood sugar varies. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

A Review of Nateglinide in the Management of Type 2 Diabetes
8 ISSN: 2347-7881 Review Article A Review of Nateglinide in the Management of Type 2 Diabetes Mistry Ripal*, Haresh T. Mulani Department of Pharmaceutics Indubhai Patel College of Pharmacy and Research Centre, Dharmaj, Gujarat, India *[email protected] ABSTRACT Diabetes mellitus, or simply diabetes, is a chronic disease that occurs when the pancreas is no longer able to make insulin, or when the body cannot make good use of the insulin it produces. The chronic hyperglycemia of diabetes is associated with long-term damage, dysfunction, and failure of different organs, especially the eyes, kidneys, nerves, heart, and blood vessels. The success of oral hypoglycemic drug therapy is usually based on are storation of normal blood glucose levels. Traditionally, the term oral hypoglycemic was used interchangeably with sulfonylureas, but more recently the development of several new drugs has broadened this designation to include all oral medications for diabetes. Nateglinide, a D-phenylalanine derivative, is a novel insulinotropic drug which has recently been launched as a therapeutic agent for Type II diabetes. Clinical studies have shown that nateglinide induces a rapid onset of insulin release synergistic with meal administration which effectively restores the early phase of insulin secretion. Since the loss of early insulin secretion is thought to play an important role in the development of glucose in tolerance the ability of nateglinide to promote early insulin release is potentially of considerable therapeutic benefit. Keywords: Nateglinide, diabetes mellitus, Glucose Homeostasis INTRODUCTION diabetics in the country. [2] The recent rise in Vast amounts of literature have been written obesity in the United States accounts for much about the current and growing epidemic of of the observed and anticipated rise in cases of diabetes mellitus (DM) and its associated diabetes mellitus in this country. -
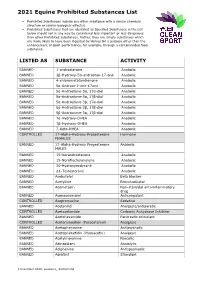
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

Association Between Use of Warfarin with Common
RESEARCH BMJ: first published as 10.1136/bmj.h6223 on 7 December 2015. Downloaded from OPEN ACCESS Association between use of warfarin with common sulfonylureas and serious hypoglycemic events: retrospective cohort analysis John A Romley,1,2 Cynthia Gong,3 Anupam B Jena,4 Dana P Goldman,1,5,4 Bradley Williams,6 Anne Peters7 1Leonard D. Schaeffer Center for ABSTRACT consciousness/mental status (2490/416 479 v Health Policy and Economics STUDY QUESTION 14 414/3 938 939; adjusted odds ratio 1.22, 1.16 to 2Price School of Public Policy, Is warfarin use associated with an increased risk of 1.29). Unmeasured factors could be correlated with University of Southern serious hypoglycemic events among older people both warfarin use and serious hypoglycemic events, California, 635 Downey Way, Los Angeles, CA 90089-3333, treated with the sulfonylureas glipizide and leading to confounding. The findings may not USA glimepiride? generalize beyond the elderly Medicare population. 3 School of Pharmacy, University METHODS WHAT THIS STUDY ADDS of Southern California, 635 Downey Way, Los Angeles, CA This was a retrospective cohort analysis of pharmacy A substantial positive association was seen between 90089-3333, USA and medical claims from a 20% random sample of use of warfarin with glipizide/glimepiride and hospital 4Department of Health Care Medicare fee for service beneficiaries aged 65 years or admission/emergency department visits for Policy, Harvard Medical School, older. It included 465 918 beneficiaries with diabetes hypoglycemia and related diagnoses, particularly in 180 Longwood Avenue, Boston, MA 02115, USA who filled a prescription for glipizide or glimepiride patients starting warfarin. -

General Practice Prescribing Trends in England and Wales – Annual Review 2014
General Practice Prescribing 2016 Trends General Practice Prescribing Trends in England and Wales Annual Review 2016 cogora.com 978-1-911392-02-6 Contents Report authors: About Cogora 04 Ejike Nwokoro, MD. Abbreviations 04 Senior Analyst, Insight & Market Access, Cogora Executive summary 05 Ellen Murphy, PhD. Methodology 07 Head of Insight & Market Access, Cogora Introduction 08 Diabetes 13 Chantal Hinds, PhD. Respiratory Corticosteroids 16 Interim Head of Insight & Market Access, Cogora Analgesics 20 Victoria Stanway, MSc. Antiepileptics 25 Senior Analyst, Insight & Market Access, Cogora Oral Nutrition 30 Mahmoud El Ghannam, MSc. References 34 Analyst, Insight & Market Access, Cogora Appendix 36 Copyright © Cogora 2018 The contents of this publication are protected by copyright. All rights reserved. The contents of this publication, either in whole or in part, may not be reproduced, stored in a data retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without written permission of the publisher. Action will be taken against companies or individual persons who ignore this warning. The information set forth herein has been obtained from sources which we believe to be reliable but is not guaranteed. This publication is provided with the understanding that the authors and publisher shall have no liability for any errors, inaccuracies or omissions therein and, by this publication, the authors and publisher are not engaged in rendering consulting advice or other professional advice to the recipient with regard to any specific matter. In the event that consulting or other expert assistance is required with regard to any specific matter, the services of qualified professionals should be sought. -

Cyclohexanes in Drug Discovery
Cyclohexanes in Drug Discovery Overview The cyclohexyl fragment is a popular building in both natural and Key Points synthetic drugs, serving as either the core structure or as part of a peripheral side chain. The cyclohexyl group may function as a May function as a bioisostere for the t-butyl group for a bioisostere for the t-butyl group for a deeper hydrophilic pocket on target protein. As a rigid version of floppy alkyl chain, the deeper hydrophilic pocket on target protein . cyclohexyl replacement reduces entropy and may offer better M ay offer better affinity. affinity. As a bioisotere for the flat phenyl group, cyclohexyl May offer more contact points substituent has the advantage of being three dimensional, which with target protein. potentially offers more contact points with target protein. This concept has been proven in the discovery of venetoclax (Venclexta). In addition, the cyclohexenyl motif is a metabolically more stable bioisostere for furanose and this concept has been demonstrated by the success of oseltamivir (Tamiflu). Bridging Molecules for Innovative Medicines 1 PharmaBlock designs and As a bioisotere for the t-butyl moiety, cyclohexyl fragment occupies more synthesizes over 2472 space, which could be beneficial when binding to a deeper lipophilic pocket Cyclohexanes and 358 on the target protein. On the other hand, as a bioisotere for the flat phenyl Cyclohexyl products are in group, cyclohexyl substituent has the advantage of being three stock. CLICK HERE to find dimensional, which potentially offers more contact points with target detailed product protein. information on webpage. Cyclohexane-containing Drugs Many drugs isolated from Nature contain the cyclohexyl group either as the core structure such as in dihydroartemisinin (1) or on the side-chain such as in the case of sirolimus (rapamycin, Rapamune, 2). -

OHA) in Complex Clinical Situations and Important Drug Interactions with OHA’S
Management guidelines for use of oral hypoglycemic agents (OHA) in complex clinical situations and important drug interactions with OHA’s J. S. Saini ABSTRACT Renal diseases result in the decreased renal elimination of sulfonylureas and their active Special precautions need to be exercised with the metabolites, thus prolonging their action. Therefore use of oral hypoglycemic drugs (OHA) in complex in renal failure, hypoglycemia is likely to develop clinical situations. These precautions are based on regardless of the drugs used (4, 5). The half-life of the pharmacologic properties of the drugs in the all sulfonylureas is prolonged in patients with renal normal and disease state. We describe the use of disease. This is of little clinical importance as far as OHA in hepatic and renal diseases. Drug tolbutamide and glipizide are concerned, since only interactions of OHA and lactic acidosis due to small amounts are excreted intact in the urine. biguanides is also described. Acetohexamide, on the other hand has more active metabolites (hydroxyhexamide) which can 1. USE OF OHA IN HEPATIC DISEASES: accumulate in renal insufficiency and can produce pronounced hypoglycemia (6). Chlorpropamide, Information regarding effects of OHA in hepatic because of its long half-life, also should not be used dysfunction is meagre (1, 2). Usually, hepatic in patients with renal disease. Tolazamide and metabolism of sulfonylurea involves inactivation by glibenclamide have active metabolites that a process of oxidation rather than conjugation. accumulate in patients with severe renal Conjugation involves either a glucuronidation or insufficiency (with creatinine clearance less than 30 acetylation. This occurs only in case of ml/mt (1).