Gene Set Enrichment Analysis Using Single Nucleotide Polymorphisms to Identify Genes Associated with Residual Feed Intake in Cattle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Variation in Protein Coding Genes Identifies Information
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number. -

Thermal Manipulation of the Chicken Embryo Triggers Differential Gene
Loyau et al. BMC Genomics (2016) 17:329 DOI 10.1186/s12864-016-2661-y RESEARCH ARTICLE Open Access Thermal manipulation of the chicken embryo triggers differential gene expression in response to a later heat challenge Thomas Loyau1, Christelle Hennequet-Antier1, Vincent Coustham1, Cécile Berri1, Marie Leduc1, Sabine Crochet1, Mélanie Sannier1, Michel Jacques Duclos1, Sandrine Mignon-Grasteau1, Sophie Tesseraud1, Aurélien Brionne1, Sonia Métayer-Coustard1, Marco Moroldo2, Jérôme Lecardonnel2, Patrice Martin3, Sandrine Lagarrigue4, Shlomo Yahav5 and Anne Collin1* Abstract Background: Meat type chickens have limited capacities to cope with high environmental temperatures, this sometimes leading to mortality on farms and subsequent economic losses. A strategy to alleviate this problem is to enhance adaptive capacities to face heat exposure using thermal manipulation (TM) during embryogenesis. This strategy was shown to improve thermotolerance during their life span. The aim of this study was to determine the effects of TM (39.5 °C, 12 h/24 vs 37.8 °C from d7 to d16 of embryogenesis) and of a subsequent heat challenge (32 °C for 5 h) applied on d34 on gene expression in the Pectoralis major muscle (PM). A chicken gene expression microarray (8 × 60 K) was used to compare muscle gene expression profiles of Control (C characterized by relatively high body temperatures, Tb) and TM chickens (characterized by a relatively low Tb) reared at 21 °C and at 32 °C (CHC and TMHC, respectively) in a dye-swap design with four comparisons and 8 broilers per treatment. Real-time quantitative PCR (RT-qPCR) was subsequently performed to validate differential expression in each comparison. -
Primepcr™Assay Validation Report
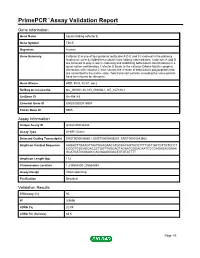
PrimePCR™Assay Validation Report Gene Information Gene Name tubulin folding cofactor E Gene Symbol TBCE Organism Human Gene Summary Cofactor E is one of four proteins (cofactors A D E and C) involved in the pathway leading to correctly folded beta-tubulin from folding intermediates. Cofactors A and D are believed to play a role in capturing and stabilizing beta-tubulin intermediates in a quasi-native confirmation. Cofactor E binds to the cofactor D/beta-tubulin complex; interaction with cofactor C then causes the release of beta-tubulin polypeptides that are committed to the native state. Two transcript variants encoding the same protein have been found for this gene. Gene Aliases HRD, KCS, KCS1, pac2 RefSeq Accession No. NC_000001.10, NG_009230.1, NT_167186.1 UniGene ID Hs.498143 Ensembl Gene ID ENSG00000116957 Entrez Gene ID 6905 Assay Information Unique Assay ID qHsaCID0036444 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000366601, ENST00000406207, ENST00000543662 Amplicon Context Sequence AAGAGTTGAAGTTAATGGAGAACATGCAACAGTACGTTTTGCTGGTGTTGTCCCT CCCGTGGCAGGACCCTGGTTAGGAGTAGAATGGGACAATCCCGAGAGAGGAAA GCATGATGGGAGCCACGAAGGGACTGTGTATTTT Amplicon Length (bp) 112 Chromosome Location 1:235543400-235564894 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 90 R2 0.9896 cDNA Cq 20.95 cDNA Tm (Celsius) 83.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 33.46 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation -

Species Groups Distributed Across Elevational Gradients Reveal Convergent and Continuous Genetic Adaptation to High Elevations
Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations Yan-Bo Suna,1, Ting-Ting Fua,b,1, Jie-Qiong Jina, Robert W. Murphya,c, David M. Hillisd,2, Ya-Ping Zhanga,e,f,2, and Jing Chea,e,g,2 aState Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, 650223 Kunming, China; bKunming College of Life Science, University of Chinese Academy of Sciences, 650204 Kunming, China; cCentre for Biodiversity and Conservation Biology, Royal Ontario Museum, Toronto, ON M5S 2C6, Canada; dDepartment of Integrative Biology and Biodiversity Center, University of Texas at Austin, Austin, TX 78712; eCenter for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, 650223 Kunming, China; fState Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, 650091 Kunming, China; and gSoutheast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, 05282 Nay Pyi Taw, Myanmar Contributed by David M. Hillis, September 7, 2018 (sent for review August 7, 2018; reviewed by John H. Malone and Fuwen Wei) Although many cases of genetic adaptations to high elevations Most previous studies of the genetic processes of HEA have have been reported, the processes driving these modifications and compared species or populations from high elevations above the pace of their evolution remain unclear. Many high-elevation 3,500 m with those from low elevations to identify sequence adaptations (HEAs) are thought to have arisen in situ as popula- variation and/or expression shifts in the high-elevation group (8– tions rose with growing mountains. -

Drosophila Tubulin-Specific Chaperone E Functions At
Page nos Page total Colour pages: Facing pages: Issue Ms order DEVELOPMENT AND DISEASE RESEARCH ARTICLE 1 Development 136, 0000-0000 (2009) doi:10.1242/dev.029983 Drosophila Tubulin-specific chaperone E functions at neuromuscular synapses and is required for microtubule network formation Shan Jin1,2, Luyuan Pan1, Zhihua Liu1, Qifu Wang1, Zhiheng Xu1 and Yong Q. Zhang1,* Hypoparathyroidism, mental retardation and facial dysmorphism (HRD) is a fatal developmental disease caused by mutations in tubulin-specific chaperone E (TBCE). A mouse Tbce mutation causes progressive motor neuronopathy. To dissect the functions of TBCE and the pathogenesis of HRD, we generated mutations in Drosophila tbce, and manipulated its expression in a tissue-specific manner. Drosophila tbce nulls are embryonic lethal. Tissue-specific knockdown and overexpression of tbce in neuromusculature resulted in disrupted and increased microtubules, respectively. Alterations in TBCE expression also affected neuromuscular synapses. Genetic analyses revealed an antagonistic interaction between TBCE and the microtubule-severing protein Spastin. Moreover, treatment of muscles with the microtubule-depolymerizing drug nocodazole implicated TBCE as a tubulin polymerizing protein. Taken together, our results demonstrate that TBCE is required for the normal development and function of neuromuscular synapses and that it promotes microtubule formation. As defective microtubules are implicated in many neurological and developmental diseases, our work on TBCE may offer novel insights into their basis. KEY WORDS: Drosophila, HRD, Spastin, TBCE (CG7861), Tubulin chaperone INTRODUCTION at the MT organizing center, perturbed MT polarity and decreased Microtubules (MTs), one of the major building blocks of cells, play precipitable MT, while total tubulin remains unchanged (Parvari et a crucial role in a diverse array of biological functions including cell al., 2002). -

Circulating Mir-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1
Journal of Personalized Medicine Article Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis Siyuan Huang 1, Yong-Kai Wei 2, Satyavani Kaliamurthi 3,4 , Yanghui Cao 5, Asma Sindhoo Nangraj 6, Xin Sui 1, Dan Chu 7, Huan Wang 7, Dong-Qing Wei 3,4,6 , Gilles H. Peslherbe 3, Gurudeeban Selvaraj 3,4 and Jiang Shi 7,* 1 Department of Oncology, The First Affiliated Hospital of Zhengzhou University, No.1 Jianshe East Road, Zhengzhou 450052, China; [email protected] (S.H.); [email protected] (X.S.) 2 College of Science, Henan University of Technology, Zhengzhou 450001, China; [email protected] 3 Centre for Research in Molecular Modeling and Department of Chemistry and Biochemistry, Concordia University, 7141 Sherbrooke Street West, Montréal, QC H4B 1R6, Canada; [email protected] (S.K.); [email protected] (D.-Q.W.); [email protected] (G.H.P.); [email protected] (G.S.) 4 Center of Interdisciplinary Science-Computational Life Sciences, College of Biological Engineering, Henan University of Technology, No.100, Lianhua Street, Hi-Tech Development Zone, Zhengzhou 450001, China 5 Department of General Surgery, Henan Tumor Hospital, No.127 Dongming Road, Zhengzhou 450008, China; [email protected] 6 The State Key Laboratory of Microbial Metabolism, College of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China; [email protected] 7 Department of Respiratory, The First Affiliated Hospital of Zhengzhou University, No.1 Jianshe East Road, Zhengzhou 450052, China; [email protected] (D.C.); [email protected] (H.W.) * Correspondence: [email protected]; Tel.: +86-15824836717 Received: 20 August 2020; Accepted: 28 September 2020; Published: 11 October 2020 Abstract: Analysis of circulating miRNAs (cmiRNAs) before surgical operation (BSO) and after the surgical operation (ASO) has been informative for lung adenocarcinoma (LUAD) diagnosis, progression, and outcomes of treatment. -

Microarray-Based Identification of Genes Associated with Cancer
Yin et al. Journal of Experimental & Clinical Cancer Research (2016) 35:127 DOI 10.1186/s13046-016-0403-2 RESEARCH Open Access Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma Fuqiang Yin1,2†, Lipei Shu3†, Xia Liu4†, Ting Li1, Tao Peng3, Yueli Nan5, Shu Li5, Xiaoyun Zeng2,5* and Xiaoqiang Qiu5* Abstract Background: Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths. The average survival and 5-year survival rates of HCC patients still remains poor. Thus, there is an urgent need to better understand the mechanisms of cancer progression in HCC and to identify useful biomarkers to predict prognosis. Methods: Public data portals including Oncomine, The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) profiles were used to retrieve the HCC-related microarrays and to identify potential genes contributed to cancer progression. Bioinformatics analyses including pathway enrichment, protein/gene interaction and text mining were used to explain the potential roles of the identified genes in HCC. Quantitative real-time polymerase chain reaction analysis and Western blotting were used to measure the expression of the targets. The data were analysed by SPSS 20. 0software. Results: We identified 80 genes that were significantly dysregulated in HCC according to four independent microarrays covering 386 cases of HCC and 327 normal liver tissues. Twenty genes were consistently and stably dysregulated in the four microarrays by at least 2-fold and detection of gene expression by RT-qPCR and western blotting showed consistent expression profiles in 11 HCC tissues compared with corresponding paracancerous tissues. -

Drosophila Dtbce Recruits Tubulin Around Chromatin to Promote Mitotic Spindle Assembly
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.21.912428; this version posted January 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Drosophila dTBCE recruits tubulin around chromatin to promote mitotic spindle assembly Mathieu Métivier1, Emmanuel Gallaud1, Aude Pascal1, Jean-Philippe Gagné2, Guy G. Poirier2, Denis Chrétien1, Romain Gibeaux1, Laurent Richard-Parpaillon1, Christelle Benaud1, and Régis Giet1*. 1. Univ Rennes, CNRS, IGDR (Institut de Génétique et Développement de Rennes) - UMR 6290, F-35000 Rennes, France 2. Centre de Recherche du Centre Hospitalier Universitaire de Québec - Pavillon CHUL, Université Laval, Québec, QC, Canada. *. Corresponding author: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.01.21.912428; this version posted January 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Proper assembly of mitotic spindles requires microtubule nucleation at centrosomes but also around chromatin. In this study, we reveal a novel mechanism by which an enrichment of tubulin in the nuclear space following nuclear envelope breakdown promotes nucleation of spindle microtubules. This event mediated by the tubulin-specific chaperone dTBCE, depends on its tubulin binding CAP-Gly motif and is regulated by Ran. -

Gnomad Lof Supplement
1 gnomAD supplement gnomAD supplement 1 Data processing 4 Alignment and read processing 4 Variant Calling 4 Coverage information 5 Data processing 5 Sample QC 7 Hard filters 7 Supplementary Table 1 | Sample counts before and after hard and release filters 8 Supplementary Table 2 | Counts by data type and hard filter 9 Platform imputation for exomes 9 Supplementary Table 3 | Exome platform assignments 10 Supplementary Table 4 | Confusion matrix for exome samples with Known platform labels 11 Relatedness filters 11 Supplementary Table 5 | Pair counts by degree of relatedness 12 Supplementary Table 6 | Sample counts by relatedness status 13 Population and subpopulation inference 13 Supplementary Figure 1 | Continental ancestry principal components. 14 Supplementary Table 7 | Population and subpopulation counts 16 Population- and platform-specific filters 16 Supplementary Table 8 | Summary of outliers per population and platform grouping 17 Finalizing samples in the gnomAD v2.1 release 18 Supplementary Table 9 | Sample counts by filtering stage 18 Supplementary Table 10 | Sample counts for genomes and exomes in gnomAD subsets 19 Variant QC 20 Hard filters 20 Random Forest model 20 Features 21 Supplementary Table 11 | Features used in final random forest model 21 Training 22 Supplementary Table 12 | Random forest training examples 22 Evaluation and threshold selection 22 Final variant counts 24 Supplementary Table 13 | Variant counts by filtering status 25 Comparison of whole-exome and whole-genome coverage in coding regions 25 Variant annotation 30 Frequency and context annotation 30 2 Functional annotation 31 Supplementary Table 14 | Variants observed by category in 125,748 exomes 32 Supplementary Figure 5 | Percent observed by methylation. -

Missense Mutation in the Tubulin-Specific Chaperone E (Tbce
JCBReport Missense mutation in the tubulin-specific chaperone E (Tbce) gene in the mouse mutant progressive motor neuronopathy, a model of human motoneuron disease Heike Bömmel,1 Gang Xie,1 Wilfried Rossoll,1 Stefan Wiese,1 Sibylle Jablonka,1 Thomas Boehm,2 and Michael Sendtner1 1Institute of Clinical Neurobiology, University of Würzburg, 97080 Würzburg, Germany 2Max-Planck-Institute for Immunobiology, 79108 Freiburg, Germany rogressive motor neuronopathy (pmn) mutant mice (Tbce) gene on mouse chromosome 13. The human have been widely used as a model for human moto- orthologue maps to chromosome 1q42.3. The Tbce gene P neuron disease. Mice that are homozygous for the encodes a protein (cofactor E) that is essential for the form- pmn gene defect appear healthy at birth but develop ation of primary ␣-tubulin and -tubulin heterodimeric progressive motoneuron disease, resulting in severe skeletal complexes. Isolated motoneurons from pmn mutant mice muscle weakness and respiratory failure by postnatal week exhibit shorter axons and axonal swelling with irregularly 3. The disease starts at the motor endplates, and then leads structured -tubulin and tau immunoreactivity. Thus, the to axonal loss and finally to apoptosis of the corresponding pmn gene mutation provides the first genetic evidence that cell bodies. We localized the genetic defect in pmn mice to alterations in tubulin assembly lead to retrograde degeneration a missense mutation in the tubulin-specific chaperone E of motor axons, ultimately resulting in motoneuron cell death. Introduction Autosomal recessive traits of motoneuron disease are observed well as RhoGEF in ALS2 (Hadano et al., 2001; Yang et al., in classical spinal muscular atrophy (SMA;* Lefebvre et al., 2001), has given a first hint from human genetic studies that 1995), in SMA with respiratory distress type 1 (SMARD1; signaling mechanisms regulating cytoskeletal turnover might Grohmann et al., 2001), and in a rare form of juvenile also be disturbed in motoneuron disease. -

Human Leucine-Rich Repeat Proteins: a Genome-Wide Bioinformatic Categorization and Functional Analysis in Innate Immunity
Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity Aylwin C. Y. Nga,b,1, Jason M. Eisenberga,b,1, Robert J. W. Heatha, Alan Huetta, Cory M. Robinsonc, Gerard J. Nauc, and Ramnik J. Xaviera,b,2 aCenter for Computational and Integrative Biology, and Gastrointestinal Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114; bThe Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, MA 02142; and cMicrobiology and Molecular Genetics, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261 Edited by Jeffrey I. Gordon, Washington University School of Medicine, St. Louis, MO, and approved June 11, 2010 (received for review February 17, 2010) In innate immune sensing, the detection of pathogen-associated proteins have been implicated in human diseases to date, notably molecular patterns by recognition receptors typically involve polymorphisms in NOD2 in Crohn disease (8, 9), CIITA in leucine-rich repeats (LRRs). We provide a categorization of 375 rheumatoid arthritis and multiple sclerosis (10), and TLR5 in human LRR-containing proteins, almost half of which lack other Legionnaire disease (11). identifiable functional domains. We clustered human LRR proteins Most LRR domains consist of a chain of between 2 and 45 by first assigning LRRs to LRR classes and then grouping the proteins LRRs (12). Each repeat in turn is typically 20 to 30 residues long based on these class assignments, revealing several of the resulting and can be divided into a highly conserved segment (HCS) fol- protein groups containing a large number of proteins with certain lowed by a variable segment (VS). -

Gene Co-Expression Network Analysis of Reproductive Traits in Bovine Genome
Reprod Dev Biol 37(4) : 185-192 (2013) ISSN : 1738-2432 (P r i n t) http://dx.doi.org/10.12749/RDB.2013.37.4.185 ISSN : 2288-0151 (Online) Gene Co-Expression Network Analysis of Reproductive Traits in Bovine Genome Dajeong Lim1,†, Yong-Min Cho1, Seung-Hwan Lee2, Han-Ha Chai1 and Tae-Hun Kim1 1Division of Animal Genomics and Bioinformatics, National Institute of Animal Science, RDA, Suwon 441-706, Korea 2Hanwoo Experiment Station, National Institute of Animal Science, RDA, Pyungchang, 232-950, Korea ABSTRACT Many countries have implemented genetic evaluation for fertility traits in recent years. In particular, reproductive trait is a complex trait and need to require a system-level approach for identifying candidate genes related to the trait. To find the candidate gene associated with reproductive trait, we applied a weighted gene co-expression network ana- lysis from expression value of bovine genes. We identified three co-expressed modules associated with reproductive trait from bovine microarray data. Hub genes (ZP4, FHL2 and EGR4) were determined in each module; they were topologically centered with statistically significant value in the gene co-expression network. We were able to find the highly co-expressed gene pairs with a correlation coefficient. Finally, the crucial functions of co-expressed modules were reported from functional enrichment analysis. We suggest that the network-based approach in livestock may an important method for analyzing the complex effects of candidate genes associated with economic traits like repro- duction. (Key words : Reproduction, Gene Co-expression Network (GCN), Cattle) INTRODUCTION These networks provide information on the genetic con- trol of complex traits and can help identify causal ge- nes that affect gene function rather than gene expre- A good reproductive performance is crucial for eco- ssion (Haley and de Koning, 2006).