Molecular Diversity of Visual Pigments in Stomatopoda (Crustacea)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Learning in Stomatopod Crustaceans
International Journal of Comparative Psychology, 2006, 19 , 297-317. Copyright 2006 by the International Society for Comparative Psychology Learning in Stomatopod Crustaceans Thomas W. Cronin University of Maryland Baltimore County, U.S.A. Roy L. Caldwell University of California, Berkeley, U.S.A. Justin Marshall University of Queensland, Australia The stomatopod crustaceans, or mantis shrimps, are marine predators that stalk or ambush prey and that have complex intraspecific communication behavior. Their active lifestyles, means of predation, and intricate displays all require unusual flexibility in interacting with the world around them, imply- ing a well-developed ability to learn. Stomatopods have highly evolved sensory systems, including some of the most specialized visual systems known for any animal group. Some species have been demonstrated to learn how to recognize and use novel, artificial burrows, while others are known to learn how to identify novel prey species and handle them for effective predation. Stomatopods learn the identities of individual competitors and mates, using both chemical and visual cues. Furthermore, stomatopods can be trained for psychophysical examination of their sensory abilities, including dem- onstration of color and polarization vision. These flexible and intelligent invertebrates continue to be attractive subjects for basic research on learning in animals with relatively simple nervous systems. Among the most captivating of all arthropods are the stomatopod crusta- ceans, or mantis shrimps. These marine creatures, unfamiliar to most biologists, are abundant members of shallow marine ecosystems, where they are often the dominant invertebrate predators. Their common name refers to their method of capturing prey using a folded, anterior raptorial appendage that looks superficially like the foreleg of a praying mantis. -

Linkage Mechanics and Power Amplification of the Mantis Shrimp's
3677 The Journal of Experimental Biology 210, 3677-3688 Published by The Company of Biologists 2007 doi:10.1242/jeb.006486 Linkage mechanics and power amplification of the mantis shrimp’s strike S. N. Patek1,*, B. N. Nowroozi2, J. E. Baio1, R. L. Caldwell1 and A. P. Summers2 1Department of Integrative Biology, University of California, Berkeley, CA 94720-3140, USA and 2Ecology and Evolutionary Biology, University of California–Irvine, Irvine, CA 92697-2525, USA *Author for correspondence (e-mail: [email protected]) Accepted 6 August 2007 Summary Mantis shrimp (Stomatopoda) generate extremely rapid transmission is lower than predicted by the four-bar model. and forceful predatory strikes through a suite of structural The results of the morphological, kinematic and modifications of their raptorial appendages. Here we mechanical analyses suggest a multi-faceted mechanical examine the key morphological and kinematic components system that integrates latches, linkages and lever arms and of the raptorial strike that amplify the power output of the is powered by multiple sites of cuticular energy storage. underlying muscle contractions. Morphological analyses of Through reorganization of joint architecture and joint mechanics are integrated with CT scans of asymmetric distribution of mineralized cuticle, the mantis mineralization patterns and kinematic analyses toward the shrimp’s raptorial appendage offers a remarkable example goal of understanding the mechanical basis of linkage of how structural and mechanical modifications can yield dynamics and strike performance. We test whether a four- power amplification sufficient to produce speeds and forces bar linkage mechanism amplifies rotation in this system at the outer known limits of biological systems. -

MANTIS SHRIMPSSHRIMPS Fast, Flexible, Fearless - Scurrying and Scooting Among the Coral Rubble Or Suddenly Exploding from Their Burrows in the Muck
93 Spotlight The Peacock Mantis Shrimp Odontodactylus scyllarus is - as its common name implies - the most colorful species among these widespread crustacean predators. THETHE LURKINGLURKING HORRORHORROR OFOF THETHE DEEPDEEP MANTISMANTIS SHRIMPSSHRIMPS Fast, flexible, fearless - scurrying and scooting among the coral rubble or suddenly exploding from their burrows in the muck. To impale and smash their hapless prey GOOGLE EARTH COORDINATES HERE 94 TEXT BY ANDREA FERRARI PHOTOS BY ANDREA & ANTONELLA FERRARI Wreathed any newcomers to scuba in a cloud of Mdiving are scared of sharks. Others are volcanic sand, a large afraid of morays. Some again are Lysiosquillina intimidated by barracudas... Little they sp. literally know that some of the scariest, most explodes from fearsome and probably most monstrous its burrow creatures of the deep lurk a few feet in a three- below the surface, silently waiting, millisecond coldly staring at their surroundings, attack. This is a “spearer” waiting for the opportunity to strike with species - notice a lightning-fast motion and to cruelly its sharply impale their prey or smash it to toothed smithereens! Luckily, most of these raptorial claws. terrifying critters are just a few inches long – otherwise diving on coral reefs might be a risky proposition indeed for every human being... But stop for a moment, and consider those cunning predators of the seabottom, the mantis shrimps: an elongated, segmented and armored body, capable of great flexibility and yet strong enough to resist the bite of all but the fiercest triggerfish; a series of short, parallel, jointed legs positioned under the thorax to swiftly propel it among the reefs rubble bottom; a pair of incredibly large, multifaceted dragonfly-like eyes, mounted on sophisticated swiveling joints, capable of giving the animal an absolutely unbeatable 3-D vision on a 360° field of vision, immensely better than our own and enabling it to strike with implacable accuracy at its chosen target. -

Folk Taxonomy of Marine Fauna on Takuu Atoll, Papua New Guinea
2 SPC Traditional Marine Resource Management and Knowledge Information Bulletin #39 – April 2018 Catching names: Folk taxonomy of marine fauna on Takuu Atoll, Papua New Guinea Anke Moesinger1 Abstract Folk taxonomies are a critical component for understanding resource use patterns and cultural, social and economic preferences on geographically remote Pacific atolls. To understand how people perceive and make use of their environment, 200 local names for marine vertebrates and invertebrates were collected and the hierarchical classification system was documented on Takuu Atoll in Papua New Guinea. The local nomenclature of the marine fauna of Takuu is based largely on shared fundamental morphological charac- teristics. Furthermore, all fish (Te ika) in the ocean are placed into one of five distinct groups in the hierar- chical classification system. These include three functional groups that are categorised by ecological niche, whereas another group encompasses all fish that possess a certain behavioural trait. The fifth group is unique in that it is solely made up of fish that were previously targeted during local Sii fishing expeditions. This article presents an analysis of Takuu residents’ descriptions and classifications of local fish and marine invertebrates. Keywords Folk taxonomy, Takuu Atoll, local knowledge, Polynesian outlier, folk hierarchical classification Introduction atoll is one of only three Polynesian outliers found in PNG. The others include Nukuria, also known as Takuu Atoll islanders are dependent on and inex- Fead Island, which is located 160 km to the north- tricably linked to the marine environment that sur- west of the atoll, and Nukumanu, or Tasman, which rounds them, and fishing permeates almost every is situated 315 km to the east. -

Polarization Signals in Mantis Shrimps
Polarization signals in mantis shrimps Thomas W. Cronin*a, Tsyr-Huei Chioua,b, Roy L. Caldwellc, Nicholas Robertsd, Justin Marshallb aDept. of Biological Sciences, UMBC, Baltimore, MD, USA 21250; bQBI, Univ. of Queensland, Brisbane, Australia 4072; cDept. of Integrative Biology, Univ. of California, Berkeley, CA 94720; dThe Photon Science Institute, Univ. of Manchester, Manchester, UK M13 9PL ABSTRACT While color signals are well known as a form of animal communication, a number of animals communicate using signals based on patterns of polarized light reflected from specialized body parts or structures. Mantis shrimps, a group of marine crustaceans, have evolved a great diversity of such signals, several of which are based on photonic structures. These include resonant scattering devices, structures based on layered dichroic molecules, and structures that use birefringent layers to produce circular polarization. Such biological polarizers operate in different spectral regions ranging from the near-UV to medium wavelengths of visible light. In addition to the structures that are specialized for signal production, the eyes of many species of mantis shrimp are adapted to detect linearly polarized light in the ultraviolet and in the green, using specialized sets of photoreceptors with oriented, dichroic visual pigments. Finally, a few mantis shrimp species produce biophotonic retarders within their photoreceptors that permit the detection of circularly polarized light and are thus the only animals known to sense this form of polarization. Mantis shrimps use polarized light in species-specific signals related to mating and territorial defense, and their means of manipulating light’s polarization can inspire designs for artificial polarizers and achromatic retarders. -

Ultraviolet Filters in Stomatopod Crustaceans: Diversity, Ecology and Evolution Michael J
© 2015. Published by The Company of Biologists Ltd | The Journal of Experimental Biology (2015) 218, 2055-2066 doi:10.1242/jeb.122036 RESEARCH ARTICLE Ultraviolet filters in stomatopod crustaceans: diversity, ecology and evolution Michael J. Bok*,§, Megan L. Porter‡ and Thomas W. Cronin ABSTRACT 1988; Marshall et al., 1991a) and at least five receptors sensitive to Stomatopod crustaceans employ unique ultraviolet (UV) optical filters various spectral ranges of ultraviolet (UV) light (Kleinlogel and in order to tune the spectral sensitivities of their UV-sensitive Marshall, 2009; Marshall and Oberwinkler, 1999). Underlying photoreceptors. In the stomatopod species Neogonodactylus oerstedii, these diverse visual sensitivities is an array of optical and retinal we previously found four filter types, produced by five distinct structural modifications (Horridge, 1978; Marshall et al., 1991a; mycosporine-like amino acid pigments in the crystalline cones of their Schiff et al., 1986), the expression of a great number of opsins specialized midband ommatidial facets. This UV-spectral tuning array resulting in the most visual pigments yet described in a single eye produces receptors with at least six distinct spectral sensitivities, (Cronin and Marshall, 1989b; Cronin et al., 1993; Porter et al., 2009, despite expressing only two visual pigments. Here, we present a 2013), and the tuning of spectral sensitivity via serial filtering broad survey of these UV filters across the stomatopod order, effects due to more distal visual pigments as well as photostable examining their spectral absorption properties in 21 species from colored pigments (Cronin and Marshall, 1989a; Cronin et al., seven families in four superfamilies. We found that UV filters are 1994a,b, 2014; Marshall, 1988; Marshall et al., 1991b; Porter et al., present in three of the four superfamilies, and evolutionary character 2010). -

Clam Ctenoides Ales (Finlay, 1927): Mechanisms and Behavioral Function
Flashing in the 'Disco' Clam Ctenoides ales (Finlay, 1927): Mechanisms and Behavioral Function By Lindsey Dougherty Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Integrative Biology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Roy L. Caldwell, Chair Professor David R. Lindberg Professor Damian O. Elias Spring 2016 Abstract Flashing in the 'Disco' Clam Ctenoides ales (Finlay, 1927): Mechanisms and Behavioral Function by Lindsey Dougherty Doctor of Philosophy in Integrative Biology University of California, Berkeley Professor Roy L. Caldwell, Chair This dissertation investigated the ‘disco’ clam Ctenoides ales (Limidae), which is the only bivalve in the world that has a behaviorally-mediated flashing display. Topics covered include (i) mechanisms, ultrastructure and movement that produce the flashing, (ii) the fitness value (function) of the flashing, (iii) the clams’ sensory abilities and vision, and (iv) the clams’ ecology, distribution and habitat. The flashing occurs on the clams’ mantle lip. Electron microscopy revealed two distinct tissue sides; one highly scattering side that contains dense aggregations of spheres composed of silica (white), and one highly absorbing side that does not (red). High-speed video confirmed that the two sides alternate rapidly, creating the appearance of flashing. Optical modeling suggested that the sphere’s diameter is nearly optimal for scattering light, especially at shorter wavelengths, which predominate in the ocean. This simple mechanism produces a striking optical effect. Three potential hypotheses for the fitness value of the flashing were investigated; conspecific attraction, prey luring, and/or predator deterrence. The lack of movement toward other C. -

Exceptional Diversity of Opsin Expression Patterns in Neogonodactylus Oerstedii (Stomatopoda) Retinas
Exceptional diversity of opsin expression patterns in Neogonodactylus oerstedii (Stomatopoda) retinas Megan L. Portera,b,1,2, Hiroko Awataa,1, Michael J. Boka,c, and Thomas W. Cronina aDepartment of Biological Sciences, University of Maryland Baltimore County, Baltimore, MD 21250; bDepartment of Biology, University of Hawai’iat Manoa, Honolulu, HI 96822; and cSchool of Biological Sciences, University of Bristol, BS8 1TQ Bristol, United Kingdom Edited by Claude Desplan, New York University, New York, NY, and approved February 27, 2020 (received for review October 4, 2019) Stomatopod crustaceans possess some of the most complex animal Most of the spectral diversity found in stomatopod retinas visual systems, including at least 16 spectrally distinct types of exists in the four dorsal-most ommatidial MB rows (MB rows 1 photoreceptive units (e.g., assemblages of photoreceptor cells). Here to 4). In MB rows 1 to 4, the seven proximal photoreceptor cells we fully characterize the set of opsin genes expressed in retinal tissues (R1–7) are subdivided into two separate photoreceptive units, and determine expression patterns of each in the stomatopod Neo- formed by sets of three or four retinula cells. Therefore, in MB gonodactylus oerstedii. Using a combination of transcriptome and rows 1 to 4 each ommatidium is formed from three successive RACE sequencing, we identified 33 opsin transcripts expressed in each photoreceptive layers comprising the distal UVS R8 photore- N. oerstedii eye, which are predicted to form 20 long-wavelength– ceptor, and middle and proximal photoreceptive layers sensitive sensitive, 10 middle-wavelength–sensitive, and three UV-sensitive to the visible spectrum (Fig. -
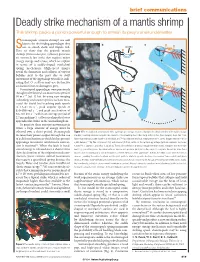
Deadly Strike Mechanism of a Mantis Shrimp This Shrimp Packs a Punch Powerful Enough to Smash Its Prey’S Shell Underwater
22.4 Brief comms MH 15/4/04 5:40 pm Page 819 brief communications Deadly strike mechanism of a mantis shrimp This shrimp packs a punch powerful enough to smash its prey’s shell underwater. tomatopods (mantis shrimp) are well known for the feeding appendages they a b c Suse to smash shells and impale fish. Saddle Here we show that the peacock mantis shrimp (Odontodactylus scyllarus) generates c an extremely fast strike that requires major v energy storage and release, which we explain 0 ms m in terms of a saddle-shaped exoskeletal spring mechanism. High-speed images p reveal the formation and collapse of vapour bubbles next to the prey due to swift d movement of the appendage towards it, indi- 1 ms cating that O. scyllarus may use destructive cavitation forces to damage its prey. Stomatopod appendages were previously thought to be limited to a maximum speed of 10msǁ1 (ref. 1) but, by using new imaging technology and a faster species,we have mea- 2 ms sured the dactyl heel reaching peak speeds of 14–23 m sǁ1,peak angular speeds of 670–990 rad sǁ1, and peak acceleration of 65–104 km sǁ2 within an average period of 2.7ms,making O.scyllarusperhaps the fastest 3 ms appendicular striker in the animal kingdom. To generate these extreme movements in water, a large amount of energy must be released over a short period. Stomatopods Figure 1 The mechanics of a stomatopod strike. a, A high-speed image sequence illustrates the distal extension of the saddle (orange increase their power output through the use triangles) occurring simultaneously with the extension of the smashing heel of the dactyl and propodus (blue triangles). -

Look at Marine Life Un Regard Sur La Vie Marine
Alook at marine life Un regard sur la vie marine A film by Jacques Perrin AND Jacques Cluzaud Alook at marine life Un regard sur la vie marine A film by Jacques Perrin AND Jacques Cluzaud Table of contents Sommaire Map of filming locations 2 Carte des lieux de tournage Mammals 6 Mammifères Birds 12 Oiseaux Reptiles 16 Reptiles Cartilaginous fishes 18 Poissons cartilagineux Bony fishes 22 Poissons osseux Molluscs 28 Mollusques Arthropods 30 Arthropodes Jellyfishes 32 Méduses Echinoderms 32 Echinodermes Tools and cameras 34 Engins et caméras Table of filmed species 38 Inventaire des espèces filmées ince The Monkey Folk in 1989, Galatée Films has forged strong epuis Le Peuple Singe en 1989, Galatée Films a tissé des liens étroits ties with the scientific community. With Winged Migration avec la communauté scientifique. AvecLe Peuple Migrateur en 2001 S in 2001, and then with ΩCEANS, the synergy between scientific D puis avec ΩCEANS, la synergie des approches scientifiques et cinémato- and cinematographic approaches was magical. Exchanges with researchers graphiques a révélé toute sa magie. Les échanges avec les chercheurs du programme from the Census of Marine Life programme have widely enhanced our Census of Marine Life ont considérablement enrichi notre perception du monde marin, marine world perception, the sensibility of our approach, and, overall, la sensibilité de notre approche, et surtout notre connaissance des créatures marines. our knowledge of marine creatures. We shot more than 500 hours of Nous avons tourné près de 500 heures de film grâce auxquelles les scientifiques pourront footage, which will enable scientists to study, as if they were right étudier, comme s’ils y étaient, la dynamique des animaux dans leur milieu sauvage. -

Flora and Fauna of North Stradbroke Island Dive Sites Australia and I Think to Myself, What a Wonderful World!
STRADDIE Flora and Fauna of North Stradbroke Island Dive Sites 104 Australia STRADDIE Flora and Fauna of North Stradbroke Island Dive Sites Australia And I think to myself, what a wonderful world! Louis Armstrong 2 3 4 Foreword North Stradbroke Island, known by Traditional Owners the Quandamooka People as Minjerribah, is internationally renowned for its spectacular scenery. From stunning white sand beaches and Pandanus-studded rocky headlands, to clear lakes and wildflower-filled bushland, the island has wonderfully diverse landscapes. Straddie is a place that once it is in your system, it is easy to love and impossible to forget. Most people can picture the crystal blue waters surrounding Straddie, but as this book highlights, that is only the beginning of the story. Sink further beneath the waves and you will find a hidden world, with some of Australia’s most prestigious dive sites. From Manta Ray Bommie – rated one of Australia’s top 10 dive sites – to Shag Rock, the diversity of underwater ecosystems echoes that found on land. This includes Flat Rock’s Shark Gutters, an important home to the endangered grey nurse shark population. Redland City Council is honoured to have partnered with The University of Queensland Underwater Club (UniDive), Sibelco Australia, Transit Systems, Manta Lodge and Scuba Centre and Point Lookout SCUBA on the Point Lookout Ecological Assessment (PLEA) of dive sites. This important project saw more than 50 UniDive volunteers conduct marine surveys on dive sites around Point Lookout. The resulting not-for-profit book not only gives a fascinating glimpse of the beauty of this underwater world, but also shines the spotlight on how crucial it is to preserve these ecological wonders. -

Devries Phd Dissertation
UC Berkeley UC Berkeley Electronic Theses and Dissertations Title The Feeding Morphology and Ecology of Stomatopod Crustaceans Permalink https://escholarship.org/uc/item/8518z4ct Author Devries, Maya Susanna Publication Date 2012 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California The Feeding Morphology and Ecology of Stomatopod Crustaceans by Maya Susanna deVries A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Integrative Biology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Sheila N. Patek, Co-Chair Professor Todd E. Dawson, Co-Chair Professor Roy L. Caldwell Professor Steven E. Beissinger Professor Peter C. Wainwright Fall 2012 The Feeding Morphology and Ecology of Stomatopod Crustaceans © 2012 by Maya Susanna deVries Abstract The Feeding Morphology and Ecology of Stomatopod Crustaceans by Maya Susanna deVries Doctor of Philosophy in Integrative Biology Professors Sheila N. Patek and Todd E. Dawson, Co-Chairs The paradigm that animals with specialized feeding morphology consume specific prey types is central to current understanding of ecological and evolutionary processes seen in nature. Mantis shrimp, or stomatopod crustaceans, are often hailed as having highly specialized feeding morphology; their raptorial appendages produce among the fastest, most powerful strikes ever reported in the animal kingdom, allowing species to capture fast-moving prey or to crush hard- shelled prey. While all stomatopods have appendages that produce fast movements, their appendage forms differ dramatically, which has led researchers to divide stomatopods into two groups: spearers that unfurl streamlined appendages to capture soft-bodied, evasive prey, and smashers that generate enough force to crush hard-shelled prey with hammer-like appendages.