New Lipase Producing Β-Proteobacteria Strains Caldimonas Sp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Response of Heterotrophic Stream Biofilm Communities to a Gradient of Resources
The following supplement accompanies the article Response of heterotrophic stream biofilm communities to a gradient of resources D. J. Van Horn1,*, R. L. Sinsabaugh1, C. D. Takacs-Vesbach1, K. R. Mitchell1,2, C. N. Dahm1 1Department of Biology, University of New Mexico, Albuquerque, New Mexico 87131, USA 2Present address: Department of Microbiology & Immunology, University of British Columbia Life Sciences Centre, Vancouver BC V6T 1Z3, Canada *Email: [email protected] Aquatic Microbial Ecology 64:149–161 (2011) Table S1. Representative sequences for each OTU, associated GenBank accession numbers, and taxonomic classifications with bootstrap values (in parentheses), generated in mothur using 14956 reference sequences from the SILVA data base Treatment Accession Sequence name SILVA taxonomy classification number Control JF695047 BF8FCONT18Fa04.b1 Bacteria(100);Proteobacteria(100);Gammaproteobacteria(100);Pseudomonadales(100);Pseudomonadaceae(100);Cellvibrio(100);unclassified; Control JF695049 BF8FCONT18Fa12.b1 Bacteria(100);Proteobacteria(100);Alphaproteobacteria(100);Rhizobiales(100);Methylocystaceae(100);uncultured(100);unclassified; Control JF695054 BF8FCONT18Fc01.b1 Bacteria(100);Planctomycetes(100);Planctomycetacia(100);Planctomycetales(100);Planctomycetaceae(100);Isosphaera(50);unclassified; Control JF695056 BF8FCONT18Fc04.b1 Bacteria(100);Proteobacteria(100);Gammaproteobacteria(100);Xanthomonadales(100);Xanthomonadaceae(100);uncultured(64);unclassified; Control JF695057 BF8FCONT18Fc06.b1 Bacteria(100);Proteobacteria(100);Betaproteobacteria(100);Burkholderiales(100);Comamonadaceae(100);Ideonella(54);unclassified; -
Additional File 1
Additional file 1 Microbial succession during the transition from active to inactive stages of deep-sea hydrothermal vent sulfide chimneys Jialin Hou, Stefan M. Sievert, Yinzhao Wang, Jeff S. Seewald, Vengadesh Perumal Natarajan Fengping Wang*, Xiang Xiao* a b 100 100 75 75 Taxa Alphaproteobacteria Taxa Betaproteobacteria Aquificae Deinococcus-Thermus Campylobacteria Deltaproteobacteria 50 Euryarchaeata 50 Euryarchaeata Others Gammaproteobacteria Percentage Percentage Thermodesulfobacterium Muproteobacteria Unclassified Nitrospirae Others Unclassified 25 25 0 0 aclA aclB cdhA cdhC napA napB norB nosZ sat soxA soxC soxY soxZ sqr aprA aprB dsrA dsrB narG nirB PRK rbcL rbcS Gene Gene Figure S1 Taxonomic classification of key functional genes retrieved from the L- and M-vent chimeny.(a) The key genes enriched in the active L-vent chimney. (b) The key genes enriched in the recently inactive M-vent chimney. Reductive bacterial type Reductive archaeal type Chlorobi group Archaeoglobus(3) Oxidative bacterial type Crenarchaeota Alphaproteobacteria(1/16) Nitrospirae(12) Acidobacteria(1/14) Betaproteobacteria Firmicutes groups Gammaproteobacteria (20) Tree scale: 1 Deltaproteobacteria(9/10) Reductive bacterial type Figure S2 Maximum-likelihood phylogeny of dsrA genes retrieved from L- and M-vent chimney. The red branches represent the dsrA genes recovered from active the L-vent chimney, while the blue ones are those from recently inactive M-vent chimney. Numbers of dsrA gene for each sample are displayed in the parenthesis after the clade name. -

Enrichment of Beneficial Cucumber Rhizosphere Microbes Mediated By
Wen et al. Horticulture Research (2020) 7:154 Horticulture Research https://doi.org/10.1038/s41438-020-00380-3 www.nature.com/hortres ARTICLE Open Access Enrichment of beneficial cucumber rhizosphere microbes mediated by organic acid secretion Tao Wen1,JunYuan1, Xiaoming He2,YueLin2,QiweiHuang1 andQirongShen 1 Abstract Resistant cultivars have played important roles in controlling Fusarium wilt disease, but the roles of rhizosphere interactions among different levels of resistant cultivars are still unknown. Here, two phenotypes of cucumber, one resistant and one with increased susceptibility to Fusarium oxysporum f.sp. cucumerinum (Foc), were grown in the soil and hydroponically, and then 16S rRNA gene sequencing and nontargeted metabolomics techniques were used to investigate rhizosphere microflora and root exudate profiles. Relatively high microbial community evenness for the Foc-susceptible cultivar was detected, and the relative abundances of Comamonadaceae and Xanthomonadaceae were higher for the Foc-susceptible cultivar than for the other cultivar. FishTaco analysis revealed that specific functional traits, such as protein synthesis and secretion, bacterial chemotaxis, and small organic acid metabolism pathways, were significantly upregulated in the rhizobacterial community of the Foc-susceptible cultivar. A machine- learning approach in conjunction with FishTaco plus metabolic pathway analysis revealed that four organic acids (citric acid, pyruvate acid, succinic acid, and fumarate) were released at higher abundance by the Foc-susceptible cultivar compared with the resistant cultivar, which may be responsible for the recruitment of Comamonadaceae, a potential beneficial microbial group. Further validation demonstrated that Comamonadaceae can be “cultured” by these organic acids. Together, compared with the resistant cultivar, the susceptible cucumber tends to assemble beneficial microbes by secreting more organic acids. -

Metagenomic Analysis of Hot Springs in Central India Reveals Hydrocarbon Degrading Thermophiles and Pathways Essential for Survival in Extreme Environments
ORIGINAL RESEARCH published: 05 January 2017 doi: 10.3389/fmicb.2016.02123 Metagenomic Analysis of Hot Springs in Central India Reveals Hydrocarbon Degrading Thermophiles and Pathways Essential for Survival in Extreme Environments Rituja Saxena 1 †, Darshan B. Dhakan 1 †, Parul Mittal 1 †, Prashant Waiker 1, Anirban Chowdhury 2, Arundhuti Ghatak 2 and Vineet K. Sharma 1* 1 Metagenomics and Systems Biology Laboratory, Department of Biological Sciences, Indian Institute of Science Education and Research, Bhopal, India, 2 Department of Earth and Environmental Sciences, Indian Institute of Science Education and Research, Bhopal, India Extreme ecosystems such as hot springs are of great interest as a source of novel extremophilic species, enzymes, metabolic functions for survival and biotechnological Edited by: products. India harbors hundreds of hot springs, the majority of which are not yet Kian Mau Goh, Universiti Teknologi Malaysia, Malaysia explored and require comprehensive studies to unravel their unknown and untapped Reviewed by: phylogenetic and functional diversity. The aim of this study was to perform a large-scale Alexandre Soares Rosado, metagenomic analysis of three major hot springs located in central India namely, Badi Federal University of Rio de Janeiro, Anhoni, Chhoti Anhoni, and Tattapani at two geographically distinct regions (Anhoni Brazil Mariana Lozada, and Tattapani), to uncover the resident microbial community and their metabolic traits. CESIMAR (CENPAT-CONICET), Samples were collected from seven distinct sites of the three hot spring locations with Argentina temperature ranging from 43.5 to 98◦C. The 16S rRNA gene amplicon sequencing *Correspondence: Vineet K. Sharma of V3 hypervariable region and shotgun metagenome sequencing uncovered a unique [email protected] taxonomic and metabolic diversity of the resident thermophilic microbial community in †These authors have contributed these hot springs. -

Microbial Community Response to Heavy and Light Crude Oil in the Great Lakes
Microbial Community Response to Heavy and Light Crude Oil in the Great Lakes Stephen Techtmann 10/24/19 Microbial Sensors Techtmann Lab @ MTU Investigating the applications of environmental microbial communities Hydraulic Fracturing Related Antibiotic Resistance Oil Bioremediation Techtmann Lab @ MTU Overview • Background on oil biodegradation • Microbial response to light and heavy crude oil in the Great Lakes • Machine learning for prediction of contamination in the Great Lakes. Oil Spills Deepwater Horizon Enbridge Line 6B Deepwater Horizon Oil Spill • 4,1000,000 bbl of oil released • Light Sweet Crude oil released • April 20, 2010 • 1101.7 miles of shoreline oiled Atlas and Hazen 2011 Enbridge Line 6B Spill – Marshall MI • 20,082 bbl of oil released • Diluted Bitumen • July 26, 2010 • 70 miles of shoreline oiled https://www.mlive.com/news/kalamazoo/2010/07/state_of_emergency_declared_as.html Oil Transmissions Pipelines in the Great Lakes Region Line 5: • 645 miles from Superior WI to Sarnia Ontario • 540,000 barrels per day • Light crude and natural gas liquids (NGLs) Crude oil Oil types and API Gravity Microbes and Biotechnology (Bioremediation) Low cost input Microbe High value output Decreased Cost Contaminant Increased Efficiency Carbon dioxide or non- toxic daughter products Carbon dioxide Microbial Biomass Petroleum Microbe Daughter Products Water Microbial Ecology and Biotechnology Low cost input Microbe High value output Decreased Cost/Increased Efficiency Complex input Input A Microbe Microbe Output A Input B Microbe Output -

REPORT on Genting Highlands Bus Crash
The Independent Advisory Panel to The Minister of Transport Malaysia REPORT on Genting Highlands Bus Crash at KM 3.6 Genting Highlands-Kuala Lumpur Road on 21 August 2013 The Independent Advisory Panel to The Minister of Transport Malaysia REPORT on Genting Highlands Bus Crash at KM 3.6 Genting Highlands-Kuala Lumpur Road on 21 August 2013 Submitted on 28 January 2014 to The Minister of Transport Malaysia 2013 Genting Highlands Bus Crash Acknowledgment The Independent Advisory Panel Members (Panel) would like to put on record special thanks to the Minister of Transport Malaysia for the appointment and trust to lead and carry out this very important task without fear or favour. As entrusted, the Panel has carried out a comprehensive evaluation and review on all investigation reports pertaining to the Genting crash and concluded with recommendations for further improvements. The Panel would also like to express its gratitude and appreciation to the following agencies for their cooperation in providing valuable information and important documents promptly to support the evaluation and review. 1. Ministry of Transport Malaysia (MOT) 2. Department of Occupational Safety and Health (DOSH) 3. Road Safety Department (RSD) 4. Genting Highlands Transport Sdn Bhd(9940-V) (GHT) 5. Genting Malaysia Berhad (58019-U) (GENM) 6. Bentong Municipal Council (BMC) 7. Public Works Department (PWD) 8. Road Transport Department (RTD) 9. PUSPAKOM Sdn Bhd (285985-U) (PUSPAKOM) 10. Land Public Transport Commission (LPTC) 11. Hospital Kuala Lumpur (HKL) Finally, the Panel is grateful to the team members of the Malaysian Institute of Road Safety Research (MIROS), in particular, the Director- General, Professor Dr. -

Anti-Bacterial Effects of Mno2 on the Enrichment of Manganese
Microbes Environ. 35(4), 2020 https://www.jstage.jst.go.jp/browse/jsme2 doi:10.1264/jsme2.ME20052 Anti-bacterial Effects of MnO2 on the Enrichment of Manganese-oxidizing Bacteria in Downflow Hanging Sponge Reactors Shuji Matsushita1,2, Takafumi Hiroe1, Hiromi Kambara1, Ahmad Shoiful1,3, Yoshiteru Aoi4, Tomonori Kindaichi1, Noriatsu Ozaki1, Hiroyuki Imachi5, and Akiyoshi Ohashi1* 1Department of Civil and Environmental Engineering, Graduate School of Advanced Science and Engineering, Hiroshima University, 1–4–1, Kagamiyama, Higashi-Hiroshima, Hiroshima 739–8527, Japan; 2Western Region Industrial Research Center, Hiroshima Prefectural Technology Research Institute, 2–10–1, Aga-minami, Kure, Hiroshima 737–0004, Japan; 3Center of Technology for the Environment, Agency for the Assessment and Application of Technology, Geostech Building, Kawasan PUSPIPTEK, Serpong, Tangerang Selatan 15314, Indonesia; 4Environmental Microbiology Laboratory, Graduate School of Advance Sciences of Matter, Hiroshima University, 2–313, Kagamiyama, Higashi-Hiroshima, Hiroshima 739–8527, Japan; and 5Department of Subsurface Geobiological Analysis and Research, Japan Agency for Marine-Earth Science & Technology, Yokosuka, Kanagawa 237–0061, Japan (Received April 26, 2020—Accepted August 2, 2020—Published online September 19, 2020) We focused on the use of abiotic MnO2 to develop reactors for enriching manganese-oxidizing bacteria (MnOB), which may then be used to treat harmful heavy metal-containing wastewater and in the recovery of useful minor metals. Downflow hanging sponge (DHS) reactors were used under aerobic and open conditions to investigate the potential for MnOB enrichment. The results of an experiment that required a continuous supply of organic feed solution containing Mn(II) demonstrated that MnOB enrichment and Mn(II) removal were unsuccessful in the DHS reactor when plain sponge cubes were used. -

Hydrogenophaga Electricum Sp. Nov., Isolated from Anodic Biofilms of an Acetate-Fed Microbial Fuel Cell
J. Gen. Appl. Microbiol., 59, 261‒266 (2013) Full Paper Hydrogenophaga electricum sp. nov., isolated from anodic biofilms of an acetate-fed microbial fuel cell Zen-ichiro Kimura and Satoshi Okabe* Division of Environmental Engineering, Faculty of Engineering, Hokkaido University, Kita-ku, Sapporo, Hokkaido 060‒8628, Japan (Received October 25, 2012; Accepted April 2, 2013) A Gram-negative, non-spore-forming, rod-shaped bacterial strain, AR20T, was isolated from an- odic biofilms of an acetate-fed microbial fuel cell in Japan and subjected to a polyphasic taxo- nomic study. Strain AR20T grew optimally at pH 7.0‒8.0 and 25°C. It contained Q-8 as the pre- dominant ubiquinone and C16:0, summed feature 3 (C16:1ω7c and/or iso-C15:02OH), and C18:1ω7c as the major fatty acids. The DNA G+C content was 67.1 mol%. A neighbor-joining phylogenetic tree revealed that strain AR20T clustered with three type strains of the genus Hydrogenophaga (H. flava, H. bisanensis and H. pseudoflava). Strain AR20T exhibited 16S rRNA gene sequence similarity values of 95.8‒97.7% to the type strains of the genus Hydrogenophaga. On the basis of phenotypic, chemotaxonomic and phylogenetic data, strain AR20T is considered a novel species of the genus Hydrogenophaga, for which the name Hydrogenophaga electricum sp. nov. is pro- posed. The type strain is AR20T (= KCTC 32195T = NBRC 109341T). Key Words—Hydrogenophaga electricum; hydrogenotrophic exoelectrogen; microbial fuel cell Introduction the MFC was analyzed. Results showed that bacteria belonging to the genera Geobacter and Hydrogenoph- Microbial fuel cells (MFCs) are devices that are able aga were abundantly present in the anodic biofilm to directly convert the chemical energy of organic community (Kimura and Okabe, 2013). -
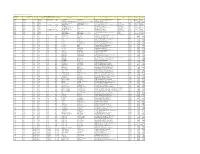
Colgate Palmolive List of Mills As of June 2018 (H1 2018) Direct
Colgate Palmolive List of Mills as of June 2018 (H1 2018) Direct Supplier Second Refiner First Refinery/Aggregator Information Load Port/ Refinery/Aggregator Address Province/ Direct Supplier Supplier Parent Company Refinery/Aggregator Name Mill Company Name Mill Name Country Latitude Longitude Location Location State AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora Agroaceite Extractora Agroaceite Finca Pensilvania Aldea Los Encuentros, Coatepeque Quetzaltenango. Coatepeque Guatemala 14°33'19.1"N 92°00'20.3"W AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora del Atlantico Guatemala Extractora del Atlantico Extractora del Atlantico km276.5, carretera al Atlantico,Aldea Champona, Morales, izabal Izabal Guatemala 15°35'29.70"N 88°32'40.70"O AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora La Francia Extractora La Francia km. 243, carretera al Atlantico,Aldea Buena Vista, Morales, izabal Izabal Guatemala 15°28'48.42"N 88°48'6.45" O Oleofinos Oleofinos Mexico Pasternak - - ASOCIACION AGROINDUSTRIAL DE PALMICULTORES DE SABA C.V.Asociacion (ASAPALSA) Agroindustrial de Palmicutores de Saba (ASAPALSA) ALDEA DE ORICA, SABA, COLON Colon HONDURAS 15.54505 -86.180154 Oleofinos Oleofinos Mexico Pasternak - - Cooperativa Agroindustrial de Productores de Palma AceiteraCoopeagropal R.L. (Coopeagropal El Robel R.L.) EL ROBLE, LAUREL, CORREDORES, PUNTARENAS, COSTA RICA Puntarenas Costa Rica 8.4358333 -82.94469444 Oleofinos Oleofinos Mexico Pasternak - - CORPORACIÓN -

1 Jejak Warisan Pahang Pencetus Tamadun
JEJAK WARISAN PAHANG PENCETUS TAMADUN PEMBANGUNAN MODAL INSAN HOLISTIK 1 Abd Jalil Borham [email protected] 1.0 Pendahuluan Menoleh ke belakang, melihat, menghimbau atau menjejaki sejarah adalah satu langkah bijak bagi memahami perkembangan lepas untuk diambil sebagai teladan dan sempadan. Membetul, melurus, dan meneguhkan segala apa yang wajar demi masa depan sesebuah bangsa dan negara. Sejarah memberi iktibar kepada masyarakat sejagat, kerana adanya hari ini membuktikan adanya semalam, dan adanya akan datang kerana adanya hari ini. Sememangnya tidak akan rugi jika sekali-kali menoleh ke belakang melihat masa lalu, merenungi zaman silam; apatah lagi melihat dan merenung untuk menimba pengalamannya. Sesebuah negara dan bangsa yang besar dan berdaulat memang mempunyai sejarah hebat bangsa dan negaranya. Namun begitu kehebatan sejarah bangsa dan negara itu tidak memberi sebarang makna dan erti jika negara dan bangsanya tidak mengambil peduli daripada pengalaman sejarah tersebut. Oleh itu sebagai bangsa yang berdaulat, tiap-tiap seorang daripadanya berkewajiban mengetahui sejarah, menghayati dan mengambil teladan serta iktibar daripada sejarah, lebih-lebih lagi sejarah negara sendiri. Melihat peri pentingnya sejarah negara ini dihayati, maka kertas kerja yang bertajuk “Jejak Warisan Negeri Pahang Darul Makmur” akan menyelusuri sejarah Pahang sejak awal dan seterusnya berusaha memasukkan Pahang dalam peta warisan dunia UNESCO. 2.0 Sejarah Pahang Sejarah Pahang bermula sebelum didirikan kerajaan Melayu Melaka, wilayah bahagian selatan Semenanjung Tanah Melayu semuanya termasuk dalam kawasan kerajaan Pahang. Orang Jawa Majapahit zaman dahulu menyebut Semenanjung Tanah Melayu sebagai Pahang sahaja. Pada awal kurun ke 16 Masihi, pada permulaan terdirinya kerajaan Melayu Johor, sempadan Pahang; di sebelahnya sampai ke Sedili Besar dan di utara sampai ke Terengganu; sempadan ke baratnya pula sampai ke Rembau, Selangor dan Perak. -

Chapter 3 Research Methodology 3.0 Study Area
27 CHAPTER 3 RESEARCH METHODOLOGY 3.0 STUDY AREA Sungai Lembing is a tin mining town 42 km northwest of Kuantan in Pahang, Malaysia. It is an old mining town, which once had the biggest tin mine on earth. The town of Sungai Lembing was once viewed as the East El Dorado of Tanah Melayu. It was once a thriving town in the late 1800’s with a population close to 30,000 consisting mainly family of miners, mining expatriates and local communities. This town was developed as a commercial centre for the local mining communities. As left abandoned in 1990, Sungai Lembing still remain the largest, longest, and deepest underground tin mine in the world. A study area for this project was located in Sg. Lembing in district of Kuantan, Pahang, East Coast of Peninsular Malaysia, about 45 km from University Malaysia Pahang (UMP) and 35 km from Kuantan town. The area was selected based on the natural factors and the condition of the river. This study area also located near to Chereh Dam, which have the biggest storage dam in Malaysia. 28 Figure 3.1: Map of the study area The characteristics of water in Sg. Lembing need to specify because it may affect the water quality of the river. The characteristics of the river depend on the more factors. Regarding to the previous research, it said there have more natural factors which affect the characteristics of the water. So, this study will determine some of characteristics at Sg. Lembing and the factors that will affect it. 3.1 Data collection The sample will be located along the Sungai Lembing from 3 stations and 2 sampling. -

Caldimonas Taiwanensis Sp. Nov., a Amylase Producing Bacterium
ARTICLE IN PRESS Systematic and Applied Microbiology 28 (2005) 415–420 www.elsevier.de/syapm Caldimonas taiwanensis sp. nov., a amylase producing bacterium isolated from a hot spring Wen-Ming Chena,Ã, Jo-Shu Changb, Ching-Hsiang Chiua, Shu-Chen Changc, Wen-Chieh Chena, Chii-Ming Jianga aDepartment of Seafood Science, National Kaohsiung Marine University, No. 142, Hai-Chuan Rd. Nan-Tzu, Kaohsiung City 811, Taiwan bDepartment of Chemical Engineering, National Cheng Kung University, Tainan, Taiwan cTajen Institute of Technology, Yen-Pu, Pingtung, Taiwan Received 22 February 2005 Abstract During screening for amylase-producing bacteria, a strain designated On1T was isolated from a hot spring located in Pingtung area, which is near the very southern part of Taiwan. Cells of this organism were Gram-negative rods motile by means of a single polar flagellum. Optimum conditions for growthwere 55 1C and pH 7. Strain On1T grew well in minimal medium containing starchas thesole carbon source, and its extracellular products expressed amylase activity. The 16S rRNA gene sequence analysis indicates that strain On1T is a member of b-Proteobacteria. On the basis of a phylogenetic analysis of 16S rDNA sequences, DNA–DNA similarity data, physiological and biochemical characteristics, as well as fatty acid compositions, the organism belonged to the genus Caldimonas and represented a novel species within this genus. The predominant cellular fatty acids of strain On1T were 16:0 (about 30%), 18:1 o7c (about 20%) and summed feature 3 (16:1o7c or 15:0 iso 2OH or both[about 31%]). Its DNA base ratio was 65.9 mol% G+C.