Table S11: Genes Identified on GISTIC on the ASCAT Algorithm
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mucosal Protection Against Sulphide: Importance of the Enzyme Rhodanese R Picton, M C Eggo, G a Merrill,Mjslangman, S Singh
201 INFLAMMATION AND INFLAMMATORY BOWEL DISEASE Gut: first published as 10.1136/gut.50.2.201 on 1 February 2002. Downloaded from Mucosal protection against sulphide: importance of the enzyme rhodanese R Picton, M C Eggo, G A Merrill,MJSLangman, S Singh ............................................................................................................................. Gut 2002;50:201–205 Background: Hydrogen sulphide (H2S) is a potent toxin normally present in the colonic lumen which may play a role in ulcerative colitis (UC). Two enzymes, thiol methyltransferase (TMT) and rhodanese (RHOD), are thought to be responsible for sulphide removal but supportive evidence is lacking. Aims: To determine the distribution of TMT and RHOD in different sites throughout the gastrointestinal tract and their efficacy as detoxifiers of H2S. Methods: Enzyme activities were measured in normal tissue resected from patients with cancer. TMT and RHOD activities were determined using their conventional substrates, 2-mercaptoethanol and sodium thiosulphate, respectively. For measurement of H S metabolism, sodium sulphide was used in See end of article for 2 authors’ affiliations the absence of dithiothreitol. Thiopurine methyltransferase (TPMT), which in common with TMT methyl- ....................... ates sulphydryl groups but is not thought to act on H2S, was also examined. Results: TMT, RHOD, and TPMT activities using their conventional substrates were found throughout Correspondence to: the gastrointestinal tract with highest activity in the colonic mucosa. When H S was given as substrate, Dr S Singh, Good Hope 2 Hospital, Sutton Coldfield, no reaction product was found with TMT or TPMT but RHOD was extremely active (Km 8.8 mM, Vmax Birmingham B75 7RR, UK; 14.6 nmol/mg/min). Incubation of colonic homogenates with a specific RHOD antibody prevented the [email protected] metabolism of H2S, indicating that RHOD is responsible for detoxifying H2S. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Rhodanese Is a Possible Enzyme Marker for Cyanide Environmental Stress on Aquatic Life
Vol. 14(29), pp. 2270-2272, 22 July, 2015 DOI: 10.5897/AJB2015.14475 Article Number: 3DC373E54256 ISSN 1684-5315 African Journal of Biotechnology Copyright © 2015 Author(s) retain the copyright of this article http://www.academicjournals.org/AJB Mini Review Rhodanese is a possible enzyme marker for cyanide environmental stress on aquatic life Okonji E. Raphael1*, Komolafe O. Olaniyi2, Fagbohunka S. Bamidele3 and Adeoye O. Akinwunmi4 1Department of Biochemistry, Obafemi Awolowo University, Ile-Ife, Nigeria. 2Department of Zoology, Obafemi Awolowo University, Ile-Ife, Nigeria. 3Department of Biochemistry, College of Basic Medicine, Olabisi Onabanjo University, Ago Iwoye, Nigeria. 4Department of Biochemistry, Federal University Oye-Ekiti, Nigeria. Received 5 February, 2015; Accepted 15 July, 2015 Rhodanese is a cyanide detoxifying enzyme. The role of man through his anthropogenic activities in and around water bodies have increased in recent times. These have led to constant exposure of water body to cyanide and cyanide compounds with increase to loss of many aquatic lives. There are limited methods employed in quick detection of cyanide in water. The aim of this paper was to present rhodanese, an enzyme, as a possible marker for detecting and monitoring water pollution as a result of environmental stress from anthropogenic activities and constant climatic changes. Key words: Rhodanese, aquatic life, water body, cyanide, toxicity. INTRODUCTION Fish and aquatic invertebrates are particularly sensitive to ecological status. It has also been reported that increase cyanide exposure. Free cyanide was reported to be the flow of water will lead to changes in sediment loads that primary toxic agent in the aquatic environment. -

Shared Sulfur Mobilization Routes for Trna Thiolation and Molybdenum Cofactor Biosynthesis in Prokaryotes and Eukaryotes
biomolecules Review Shared Sulfur Mobilization Routes for tRNA Thiolation and Molybdenum Cofactor Biosynthesis in Prokaryotes and Eukaryotes Silke Leimkühler *, Martin Bühning and Lena Beilschmidt Department of Molecular Enzymology, Institute of Biochemistry and Biology, University of Potsdam, 14476 Potsdam, Germany; [email protected] (M.B.); [email protected] (L.B.) * Correspondence: [email protected]; Tel.: +49-331-977-5603 Academic Editor: Valérie de Crécy-Lagard Received: 8 December 2016; Accepted: 9 January 2017; Published: 14 January 2017 Abstract: Modifications of transfer RNA (tRNA) have been shown to play critical roles in the biogenesis, metabolism, structural stability and function of RNA molecules, and the specific modifications of nucleobases with sulfur atoms in tRNA are present in pro- and eukaryotes. Here, especially the thiomodifications xm5s2U at the wobble position 34 in tRNAs for Lys, Gln and Glu, were suggested to have an important role during the translation process by ensuring accurate deciphering of the genetic code and by stabilization of the tRNA structure. The trafficking and delivery of sulfur nucleosides is a complex process carried out by sulfur relay systems involving numerous proteins, which not only deliver sulfur to the specific tRNAs but also to other sulfur-containing molecules including iron–sulfur clusters, thiamin, biotin, lipoic acid and molybdopterin (MPT). Among the biosynthesis of these sulfur-containing molecules, the biosynthesis of the molybdenum cofactor (Moco) and the synthesis of thio-modified tRNAs in particular show a surprising link by sharing protein components for sulfur mobilization in pro- and eukaryotes. Keywords: tRNA; molybdenum cofactor; persulfide; thiocarboxylate; thionucleosides; sulfurtransferase; L-cysteine desulfurase 1. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
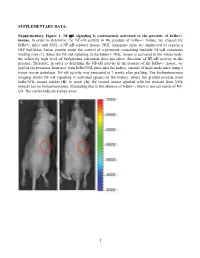
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Structural Enzymology of Sulfide Oxidation by Persulfide Dioxygenase and Rhodanese
Structural Enzymology of Sulfide Oxidation by Persulfide Dioxygenase and Rhodanese by Nicole A. Motl A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry) in the University of Michigan 2017 Doctoral Committee Professor Ruma Banerjee, Chair Assistant Professor Uhn-Soo Cho Professor Nicolai Lehnert Professor Stephen W. Ragsdale Professor Janet L. Smith Nicole A. Motl [email protected] ORCID iD: 0000-0001-6009-2988 © Nicole A. Motl 2017 ACKNOWLEDGEMENTS I would like to take this opportunity to acknowledge the many people who have provided me with guidance and support during my doctoral studies. First I would like to express my appreciation and gratitude to my advisor Dr. Ruma Banerjee for the mentorship, guidance, support and encouragement she has provided. I would like to thank my committee members Dr. Uhn-Soo Cho, Dr. Nicolai Lehnert, Dr. Stephen Ragsdale and Dr. Janet Smith for their advice, assistance and support. I would like to thank Dr. Janet Smith and members of Dr. Smith’s lab, especially Meredith Skiba, for sharing their expertise in crystallography. I would like to thank Dr. Omer Kabil for his help, suggestions and discussions in various aspects of my study. I would also like to thank members of Dr. Banerjee’s lab for their suggestions and discussions. Additionally, I would like to thank my friends and family for their support. ii TABLE OF CONTENTS ACKNOWLEDGEMENTS ii LIST OF TABLES viii LIST OF FIGURES ix ABBREVIATIONS xi ABSTRACT xii CHAPTER I. Introduction: -

Functional Investigation of Bacillus Subtilis Yrkf's Involvement in Sulfur
Peptidomics 2015; 2: 45–51 Original Study Open Access Hannah L. Martin, Katherine A. Black, Patricia C. Dos Santos* Functional investigation of Bacillus subtilis YrkF’s involvement in sulfur transfer reactions DOI 10.1515/ped-2015-0008 enzymes involved in the synthesis of the pterin organic Received August 6, 2015; accepted December 5, 2015 moiety, the insertion of sulfur and Mo to form Moco, Abstract: Sulfur incorporation into the molybdenum and the activation of their respective enzymatic partners cofactor (Moco) in the Gram-negative bacterium [4]. While components in its biosynthetic pathway vary Escherichia coli involves six enzymes. The initial between species, the structure and functionality of the reaction includes the cysteine desulfurase IscS, the active cofactor is widely conserved [5]. sulfurtransferase TusA, and the rhodanese domain- The synthesis of Moco in the Gram-negative containing protein YnjE. The Gram-positive bacterium bacterium Escherichia coli is well understood [4]. The Bacillus subtilis contains no direct homologs for IscS, organic scaffolding is synthesized from guanosine but rather four distinct cysteine desulfurases (YrvO, NifS, triphosphate (GTP) via the intermediates cyclic NifZ, SufS) and YrkF, a two-domain rhodanese protein pyranopterin monophosphate (cPMP) and molybdopterin with an N-terminal domain similar to TusA. Bioinformatic (MPT) [6], which is subsequently adenylated prior to analysis was used to identify potential enzymes involved coordinating Mo [7,8]. The conversion of cPMP to MPT in the B. subtilis Moco thiolation pathway and in vitro requires the insertion of two sulfur atoms [9], constituting reactions demonstrated that YrkF can accept sulfur from the thiolation branch of Moco biosynthesis (Figure 1). -

The Requirement of Inorganic Fe-S Clusters for the Biosynthesis of the Organometallic Molybdenum Cofactor
inorganics Review The Requirement of Inorganic Fe-S Clusters for the Biosynthesis of the Organometallic Molybdenum Cofactor Ralf R. Mendel 1, Thomas W. Hercher 1, Arkadiusz Zupok 2, Muhammad A. Hasnat 2 and Silke Leimkühler 2,* 1 Institute of Plant Biology, Braunschweig University of Technology, Humboldtstr. 1, 38106 Braunschweig, Germany; [email protected] (R.R.M.); [email protected] (T.W.H.) 2 Department of Molecular Enzymology, Institute of Biochemistry and Biology, University of Potsdam, Karl-Liebknecht-Str. 24-25, 14476 Potsdam, Germany; [email protected] (A.Z.); [email protected] (M.A.H.) * Correspondence: [email protected]; Tel.: +49-331-977-5603; Fax: +49-331-977-5128 Received: 18 June 2020; Accepted: 14 July 2020; Published: 16 July 2020 Abstract: Iron-sulfur (Fe-S) clusters are essential protein cofactors. In enzymes, they are present either in the rhombic [2Fe-2S] or the cubic [4Fe-4S] form, where they are involved in catalysis and electron transfer and in the biosynthesis of metal-containing prosthetic groups like the molybdenum cofactor (Moco). Here, we give an overview of the assembly of Fe-S clusters in bacteria and humans and present their connection to the Moco biosynthesis pathway. In all organisms, Fe-S cluster assembly starts with the abstraction of sulfur from l-cysteine and its transfer to a scaffold protein. After formation, Fe-S clusters are transferred to carrier proteins that insert them into recipient apo-proteins. In eukaryotes like humans and plants, Fe-S cluster assembly takes place both in mitochondria and in the cytosol. -

Survey of the Photosynthetic Bacteria for Rhodanese (Thiosulfate: Cyanide Sulfur Transferase) Activity DUANE C
JOURNAL OF BACTERIOLOGY, May 1971, p. 700-701 Vol. 106, No. 2 Copyright ( 1971 American Society for Microbiology Printed in U.S.A. Survey of the Photosynthetic Bacteria for Rhodanese (Thiosulfate: Cyanide Sulfur Transferase) Activity DUANE C. YOCH1 AND E. S. LINDSTROM Department of Microbiology, The Pennsylvania State University, University Park, Pennsylvania 16802 Received for publication 21 January 1971 Rhodanese activity was demonstrated in extracts from all three taxonomic fami-L: lies of photosynthetic bacteria, and this activity appeared to be uncorrelated witki-. thiosulfate metabolism. Rhodanese (thiosulfate: cyanide sulfur trans- TABLE 1. Rhodanese activity in photosynthetic ferase, EC 2.8.1.1) catalyzes the cleavage of bacteria thiosulfate (equation 1). Specific activity' CN- + S20 2- CNS- + S32- (I) Crude Solu- Chro- ble mato- This enzyme is widely distributed, having been Taxonomic Source of ex- ex- in liver and bacteria tract tract phores detected tissue, plant roots, Taxonomiy Sourac (super- (super- (pel- (4). In photosynthetic bacteria, rhodanese ac- natant natant let tivity was first demonstrated in Chromatium (5) 30,000 105,000105,000 and was later found in Rhodopseudomonas sphe- xg) xg) xg) rubrum Rho- roides and Rhodospirillum (6). Athio- Rhodospirtilum 88 171 9 danese may play a role in thiosulfate oxidation in rhodaceae rubrum Chromatium by cleaving the sulfur-sulfur bond Rhodopseudomonas 68 120 0 of thiosulfate to yield sulfur and sulfite (6). capsulata The rhodanese activity of the three taxonomic R. palustris 185 350 12 families of photosynthetic bacteria was surveyed Thio- Chromatium sp. 107 170 9 to determine if its occurrence is consistent with rhodaceae Ectothiorhodospira 116 117 8 its proposed role in thiosulfate oxidation. -

Evidence for the Physiological Role of a Rhodanese-Like Protein for the Biosynthesis of the Molybdenum Cofactor in Humans
Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans Andreas Matthies*, K. V. Rajagopalan†, Ralf R. Mendel*, and Silke Leimku¨ hler*‡ *Department of Plant Biology, Technical University Braunschweig, 38023 Braunschweig, Germany; and †Department of Biochemistry, Duke University Medical Center, Durham, NC 27710 Edited by Rowena G. Matthews, University of Michigan, Ann Arbor, MI, and approved March 3, 2004 (received for review December 10, 2003) Recent studies have identified the human genes involved in the contrast, the reaction mechanism of resulfuration of E. coli MPT biosynthesis of the molybdenum cofactor. The human MOCS3 synthase has been described in detail (8–11). Similar to ubiquitin- protein contains an N-terminal domain similar to the Escherichia activating enzymes (E1), E. coli MoeB, the MPT synthase sulfurase, coli MoeB protein and a C-terminal segment displaying similarities activates the C terminus of MoaD to form an acyl adenylate. to the sulfurtransferase rhodanese. The MOCS3 protein is believed Subsequently the MoaD acyl adenylate is converted to a thiocar- to catalyze both the adenylation and the subsequent generation of boxylate by action of any of several NifS-like proteins using L- a thiocarboxylate group at the C terminus of the smaller subunit of cysteine as the sulfur source. Sequence alignments of the human molybdopterin (MPT) synthase. The MOCS3 rhodanese-like domain MoeB homologue MOCS3 showed that the N-terminal domain is (MOCS3-RLD) was purified after heterologous expression in E. coli homologous to E. coli MoeB, but an additional C-terminal domain and was shown to catalyze the transfer of sulfur from thiosulfate is present in MOCS3 with homologies to rhodaneses (3). -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4.