Phylogenetic Relationships Between Sedum L. and Related Genera (Crassulaceae) Based on ITS Rdna Sequence Comparisons
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
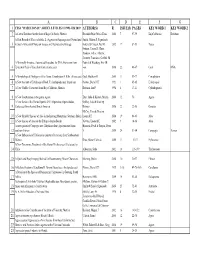
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

H3.3 Macaronesian Inland Cliff
European Red List of Habitats - Screes Habitat Group H3.3 Macaronesian inland cliff Summary The perennial vegetation of crevices and ledges of cliff faces in Macaronesia away from coastal salt-spray is of very diverse character - some, for example, dominated by succulents, others rich in ferns and bryophytes characteristic of shaded situations - and it includes several hundreds of taxa endemic to the archipelagoes. The main threats are mountaineering and rock climbing, outdoor sports and leisure activities, and construction of infrastructures such as roads and motorways. Synthesis There is no evidence of significant past reductions, either in the last 50 years or historically, and also future prospects are good, as no serious threats are envisaged, besides touristic/leisure activities and putative faulty environmental impact assessments that may overlook this habitat as valuable. Reduction in quantity, reduction in quality and criteria of geographic distribution yield the Least Concern (LC) category. In spite of the LC category, conservation policy and management should restrict to the maximum any threat to or reduction of the habitat, as it has a very high conservation value, because of high endemism of species and communities with many local and regional variaties. Overall Category & Criteria EU 28 EU 28+ Red List Category Red List Criteria Red List Category Red List Criteria Least Concern - Least Concern - Sub-habitat types that may require further examination Four general subtypes can be distinguished based on species composition and different ecological conditions. However, at present, no data are available to carry out an individual assessment of each of them. In the future, if detailed plot sampling has been carried out, such an evaluation may be possible. -

A Numerical Taxonomy of the Genus Rosularia (Dc.) Stapf from Pakistan and Kashmir
Pak. J. Bot., 44(1): 349-354, 2012. A NUMERICAL TAXONOMY OF THE GENUS ROSULARIA (DC.) STAPF FROM PAKISTAN AND KASHMIR GHULAM RASOOL SARWAR* AND MUHAMMAD QAISER Centre for Plant Conservation, University of Karachi, Karachi-75270, Pakistan Federal Urdu University of Arts, Science and Technology, Gulshan-e-Iqbal, Karachi, Pakistan Abstract Numerical analysis of the taxa belonging to the genus Rosularia (DC.) Stapf was carried out to find out their phenetic relationship. Data from different disciplines viz. general, pollen and seed morphology, chemistry and distribution pattern were used. As a result of cluster analysis two distinct groups are formed. Out of which one group consists of R. sedoides (Decne.) H. Ohba and R. alpestris A. Boriss. while other group comprises R. adenotricha (Wall. ex Edgew.) Jansson ssp. adenotricha , R. adenotricha ssp. chitralica, G.R. Sarwar, R. rosulata (Edgew.) H. Ohba and R. viguieri (Raym.-Hamet ex Frod.) G.R. Sarwar. Distribution maps of all the taxa, along with key to the taxa are also presented. Introduction studied the genus Rosularia and indicated that the genus is polyphyletic. Mayuzumi & Ohba (2004) analyzed the Rosularia is a small genus composed of 28 species, relationships within the genus Rosularia. According to distributed in arid or semiarid regions ranging from N. different workers Rosularia is polyphyletic. Africa to C. Asia through E. Mediterranean (Mabberley, There are no reports on numerical studies of 2008). Some of the taxa of Rosularia are in general Crassulaceae except the genus Sedum from Pakistan cultivation and several have great appeal due to their (Sarwar & Qaiser, 2011). The primary aim of this study is extraordinarily regular rosettes on the leaf colouring in to analyze diagnostic value of morphological characters in various seasons. -

Seeking Criteria for Biodiversity Roofs Under Finnish Conditions
SEEKING CRITERIA FOR BIODIVERSITY ROOFS UNDER FINNISH CONDITIONS WENFEI LIAO UNIVERSITY OF HELSINKI Department of Environmental Sciences Master’s Degree Programme in Environmental Ecology MASTER’S THESIS Octorber 2016 Tiedekunta/Osasto – Fakultet/Sektion – Faculty Laitos – Institution – Department Faculty of Biological and Environmental Sciences Department of Environmental Sciences Tekijä – Författare – Author Wenfei Liao Työn nimi – Arbetets titel – Title Seeking criteria for biodiversity roofs under Finnish conditions Oppiaine – Läroämne – Subject Social aspect of urban ecology Työn laji – Arbetets art – Level Aika – Datum – Month, year Sivumäärä – Sidoantal – Number of pages Master’s thesis October, 2016 76 + Appendices Tiivistelmä – Referat – Abstract Urbanisation has caused many environmental problems, such as air pollution and the loss of biodiversity. One way to mitigate these problems is to expand green spaces. Roofs, as the last frontier, could be made full use of. Green roofs have become a hot topic in recent years. In this study, I investigated the ability of green roofs to support urban biodiversity by conducting a literature review, and then I sought the criteria for biodiversity roofs under Finnish conditions by interviewing ecologists. My research questions in this study were 1) What kinds of habitats could be “ideal ecosystems” to be mimicked on biodiversity roofs in Finland; 2) which plant species could exist on roofs and whether they contribute to biodiversity; 3) what kinds of substrates support the biodiversity on roofs; 4) whether green roofs support faunal diversity and what faunal taxa could exist on roofs; 5) if and how roof structural characteristics influence roof biodiversity; 6) what kinds of management are practiced on biodiversity roofs; 7) what are people’s attitudes towards or perceptions of biodiversity roofs in general. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Sedum Society Newsletter(130) Pp
Open Research Online The Open University’s repository of research publications and other research outputs Kalanchoe arborescens - a Madagascan giant Journal Item How to cite: Walker, Colin (2019). Kalanchoe arborescens - a Madagascan giant. Sedum Society Newsletter(130) pp. 81–84. For guidance on citations see FAQs. c [not recorded] https://creativecommons.org/licenses/by-nc-nd/4.0/ Version: Version of Record Copyright and Moral Rights for the articles on this site are retained by the individual authors and/or other copyright owners. For more information on Open Research Online’s data policy on reuse of materials please consult the policies page. oro.open.ac.uk NUMBER 130 SEDUM SOCIETY NEWSLETTER JULY 2019 FRONT COVER Roy Mottram kindly supplied: “The Diet” copy of this Japanese herbal which is sharp and crisp (see page 97). “I counted the plates, and this copy is complete with 200 plates, in 8 parts, bound here in 2 vols. I checked for another Sedum but none are Established April 1987, now ending our present, so Maximowicz was basing his 32nd year. S. kagamontanum on this same plate, Subscriptions run from October to the following September. Anyone requesting translating the location as Mt. Kaga and to join after June, unless there is a special citing t.40 incorrectly. The "t.43" plate request, will receive his or her first number is also wrong. It is actually t.33 of Newsletter in October. If you do not the whole work, or Vol.2 t.8. The book is receive your copy by the 10th of April, July or October, or the 15th January, then bound back to front [by Western standards] please write to the editor: Ray as in all Japanese books of the day.” RM. -

A Numerical Taxonomy of the Genus Sedum L., from Pakistan and Kashmir
Pak. J. Bot., 43(2): 753-758, 2011. A NUMERICAL TAXONOMY OF THE GENUS SEDUM L., FROM PAKISTAN AND KASHMIR GHULAM RASOOL SARWAR AND MUHAMMAD QAISER1 Centre for Plant Conservation, University of Karachi, Karachi-75270, Pakistan 1Federal Urdu University of Arts, Science and Technology, Gulshan-e-Iqbal, Karachi, Pakistan Corresponding author e-mail: [email protected] Abstract The phenetic relationship between the species of the genus Sedum L. was investigated. Data from macro and micromorphology, including pollen and seed morphology, chemistry and distribution pattern was utilized. Two distinct groups of taxa are recognized from which one group comprises S. multicaule Wall. ex Lindl., and S. hispanicum L., while other group consists of S. trullipetalum Hook. f. & Thomson S. fischeri Raym. - Hamet and S. oreades (Decne.) Raym. – Hamet. A key of the taxa is provided and distribution maps of the species are also presented. Introduction The importance of statistical methods in biological sciences can hardly be over emphasized. Many workers were of the view that numerical analysis is the comprehensive way of evaluating and analyzing the data and producing a phenetic classification (Sokal & Sneath, 1963; Mc Neil et al., 1969; Sneath & Sokal, 1973). Cronquist (1964) debated the same issue and emphasized that data collected subjectively should not become objective, when analyzed and presented objectively. Considerable work has been done on family Crassulaceae to elucidate the phylogenetic relationship. Uhl (1963) studied the phylogeny of the family Crassulaceae. Hart (1982) analyzed the relationships within the subfamily Sedoideae. Mes (1995) did the phylogenetic analysis of 22 taxa belonging to Sempervivoideae and five taxa of Sedoideae. -

<I>Monanthes Subrosulata</I>, a New Species of <I>M.</I> Sect. <I
Willdenowia 43 – 2013 25 ÁNGEL BAÑARES BAUDET1*, AURELIO ACEVEDO RODRÍGUEZ2 & ÁNGEL REBOLÉ BEAUMONT3 Monanthes subrosulata, a new species of M. sect. Sedoidea (Crassulaceae) from La Palma, Canary Islands, Spain Abstract Bañares Baudet Á., Acevedo Rodríguez A. & Rebolé Beaumont Á.: Monanthes subrosulata, a new species of M. sect. Sedoidea (Crassulaceae) from La Palma, Canary Islands, Spain. – Willdenowia 43: 25 – 31. June 2013. – Online ISSN 1868-6397; © 2013 BGBM Berlin-Dahlem. Stable URL: http://dx.doi.org/10.3372/wi.43.43103 Monanthes subrosulata (M. sect. Sedoidea) from La Palma, Canary Islands, Spain, is described as a species new to science and illustrated. Special attention is paid to the morphological characteristics that differentiate it from the other species of the section (M. anagensis and M. laxiflora), as well as from M. muralis (M. sect. Monanthes). Additional key words: Monanthes anagensis, M. laxiflora, M. muralis, taxonomy, chorology Introduction branched habit, never forming rosettes, and with their inflorescences arising from the tips of the axes. Monan After the recent treatment of the Moroccan species thes subrosulata, located in the south and south-eastern Monanthes atlantica Ball as Sedum surculosum Coss. parts of La Palma island, shares the above characters and (Mes & ’t Hart 1994; ’t Hart & Bleij 2003), the genus unequivocally belongs to M. sect. Sedoidea. The most Monanthes Haw. is confined to the Canary Islands and striking character to separate it from the other species of Ilhas Selvagens, two archipelagos that biogeographically the section is the presence of conspicuous bladder-cell comprise the Canarian Province in the Mediterranean idioblasts (papillae), especially on the leaves (usually in- Region (Rivas-Martínez 2009), previously known as part conspicuous and not protruding in M. -

9780521118088 Index.Pdf
Cambridge University Press 978-0-521-11808-8 — The Biology of Island Floras Edited by David Bramwell , Juli Caujapé-Castells Index More Information I n d e x Abutilon , 248 , 257 , 261 Alyxia , 255 Acacia , 128 , 214 , 229 , 396 Amaranthaceae, 94 , 220 , 372 Acaena , 42 Amaranthus , 94 , 98 Acalypha , 94 , 128 , 373 , 458 , 465 Amborella , 231 Acanthaceae, 200 , 207 , 368 , 370 , 377 , 429 Amborellaceae, 230 , 231 Acantholimon , 326 Ampeloziziphus , 157 Achyranthes , 255 , 483 Amphisbaena , 156 Acidocroton , 157 Amphorogyne , 232 Acnistus , 93 , 140 , 400 , 401 , 402 Anacardiaceae, 211 , 371 , 432 Acoelorraphe , 162 anacladogenesis, 213, 220 Acrocomia , 162 Anagenesis, 212 Adansonia , 427 Anagyris , 481 , 485 adaptation, 2 , 7 , 211 , 233 , 293 , 307 , 444 , 445 , 447 , Anas , 379 493 , 504 ancient lineages, 6 , 156 adaptive radiation, 4 , 67 , 75 , 76 , 79 , 212 , 220 , 234 , Andaman, 495 445 Androcymbium , 306 , 310 , 312 , 333 Adenium , 207 aneuploidy, 5 , 278 Adiantum , 346 , 400 , 401 Angiopteris , 254 Aegialitis , 326, 329 angiosperms, 5 , 7, 18 , 19 , 158 , 160 , 161 , 228, 229, Aeonium , 286 , 309 , 342 , 494 231 , 252 , 267 , 269 , 270 , 273 , 275 , 338, 339 , 340 , Aerva , 220 342 , 344 , 346 , 348 , 349, 350 , 351 , 353 , 355 , 426 AFLPs, 93 , 136 , 284 , 288 , 302 , 309 Anisocarpus , 76 Afrolimon , 327 Annona , 131 agamospermous species, 328 Antilles, 154 , 155 , 157 , 160 , 162 , 349 Agathis , 228 , 267 Antillia , 161 Ageratum , 95 Apiaceae, 103 , 291 , 298 agriculture, 154 , 164 , 234 , 244 , 398 , 400 , 433 , 436 , Apocynaceae, -

Plethora of Plants - Collections of the Botanical Garden, Faculty of Science, University of Zagreb (2): Glasshouse Succulents
NAT. CROAT. VOL. 27 No 2 407-420* ZAGREB December 31, 2018 professional paper/stručni članak – museum collections/muzejske zbirke DOI 10.20302/NC.2018.27.28 PLETHORA OF PLANTS - COLLECTIONS OF THE BOTANICAL GARDEN, FACULTY OF SCIENCE, UNIVERSITY OF ZAGREB (2): GLASSHOUSE SUCCULENTS Dubravka Sandev, Darko Mihelj & Sanja Kovačić Botanical Garden, Department of Biology, Faculty of Science, University of Zagreb, Marulićev trg 9a, HR-10000 Zagreb, Croatia (e-mail: [email protected]) Sandev, D., Mihelj, D. & Kovačić, S.: Plethora of plants – collections of the Botanical Garden, Faculty of Science, University of Zagreb (2): Glasshouse succulents. Nat. Croat. Vol. 27, No. 2, 407- 420*, 2018, Zagreb. In this paper, the plant lists of glasshouse succulents grown in the Botanical Garden from 1895 to 2017 are studied. Synonymy, nomenclature and origin of plant material were sorted. The lists of species grown in the last 122 years are constructed in such a way as to show that throughout that period at least 1423 taxa of succulent plants from 254 genera and 17 families inhabited the Garden’s cold glass- house collection. Key words: Zagreb Botanical Garden, Faculty of Science, historic plant collections, succulent col- lection Sandev, D., Mihelj, D. & Kovačić, S.: Obilje bilja – zbirke Botaničkoga vrta Prirodoslovno- matematičkog fakulteta Sveučilišta u Zagrebu (2): Stakleničke mesnatice. Nat. Croat. Vol. 27, No. 2, 407-420*, 2018, Zagreb. U ovom članku sastavljeni su popisi stakleničkih mesnatica uzgajanih u Botaničkom vrtu zagrebačkog Prirodoslovno-matematičkog fakulteta između 1895. i 2017. Uređena je sinonimka i no- menklatura te istraženo podrijetlo biljnog materijala. Rezultati pokazuju kako je tijekom 122 godine kroz zbirku mesnatica hladnog staklenika prošlo najmanje 1423 svojti iz 254 rodova i 17 porodica. -

Mountain Plants of Northeastern Utah
MOUNTAIN PLANTS OF NORTHEASTERN UTAH Original booklet and drawings by Berniece A. Andersen and Arthur H. Holmgren Revised May 1996 HG 506 FOREWORD In the original printing, the purpose of this manual was to serve as a guide for students, amateur botanists and anyone interested in the wildflowers of a rather limited geographic area. The intent was to depict and describe over 400 common, conspicuous or beautiful species. In this revision we have tried to maintain the intent and integrity of the original. Scientific names have been updated in accordance with changes in taxonomic thought since the time of the first printing. Some changes have been incorporated in order to make the manual more user-friendly for the beginner. The species are now organized primarily by floral color. We hope that these changes serve to enhance the enjoyment and usefulness of this long-popular manual. We would also like to thank Larry A. Rupp, Extension Horticulture Specialist, for critical review of the draft and for the cover photo. Linda Allen, Assistant Curator, Intermountain Herbarium Donna H. Falkenborg, Extension Editor Utah State University Extension is an affirmative action/equal employment opportunity employer and educational organization. We offer our programs to persons regardless of race, color, national origin, sex, religion, age or disability. Issued in furtherance of Cooperative Extension work, Acts of May 8 and June 30, 1914, in cooperation with the U.S. Department of Agriculture, Robert L. Gilliland, Vice-President and Director, Cooperative Extension -

Universidad Nacional Autónoma De México Facultad De Estudios
Universidad Nacional Autónoma de México Facultad de Estudios Superiores Zaragoza “LAS CRASULÁCEAS DEL VALLE DEL MEZQUITAL’’ Tesis de licenciatura que para obtener el título de Biólogo presentan: Gabriela de Jesús Espino Ortega Luis Emilio de la Cruz López Área de Botánica Director de tesis: M. en C. Balbina Vázquez Benítez. _____________________________ Marzo 2009 UNAM – Dirección General de Bibliotecas Tesis Digitales Restricciones de uso DERECHOS RESERVADOS © PROHIBIDA SU REPRODUCCIÓN TOTAL O PARCIAL Todo el material contenido en esta tesis esta protegido por la Ley Federal del Derecho de Autor (LFDA) de los Estados Unidos Mexicanos (México). El uso de imágenes, fragmentos de videos, y demás material que sea objeto de protección de los derechos de autor, será exclusivamente para fines educativos e informativos y deberá citar la fuente donde la obtuvo mencionando el autor o autores. Cualquier uso distinto como el lucro, reproducción, edición o modificación, será perseguido y sancionado por el respectivo titular de los Derechos de Autor. Agradecimientos A la UNAM por brindarme la oportunidad de desarrollarme mejor tanto en el sentido humano como en el profesional. Por hacer la diferencia en mi vida. A la FES Zaragoza y a las personas que en ella laboran, por ser mi casa de estudios. Al grupo de sinodales formado por M. en C. Ramiro Ríos Gómez, M. en C. Balbina Vázquez Benítez, M. en C. Efraín Ángeles Cervantes, M. en C. Carlos Castillejos Cruz y Florencia Becerril Cruz, por las observaciones hechas pertinentemente para la mejora de este trabajo, por los comentarios y el tiempo dedicado al mismo. Por su amistad A la Maestra Balbina con quien estaré siempre agradecida porque trabajó con mucho entusiasmo en nuestro trabajo.