Genetic Variability in the Isolates of Rhizoctonia Solani Kühn, the Causal Agent of Sheath Blight of Rice by Using RAPD Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
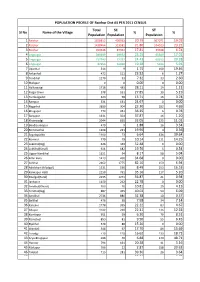
Sl No Name of the Village Total Population SC Population % ST
POPULATION PROFILE OF Raichur Dist AS PER 2011 CENSUS Total SC ST Sl No Name of the Village % % Population Population Population 1 Raichur 1928812 400933 20.79 367071 19.03 2 Raichur 1438464 313581 21.80 334023 23.22 3 Raichur 490348 87352 17.81 33048 6.74 4 Lingsugur 385699 89692 23.25 65589 17.01 5 Lingsugur 297743 72732 24.43 60393 20.28 6 Lingsugur 87956 16960 19.28 5196 5.91 7 Upanhal 514 9 1.75 100 19.46 8 Ankanhal 472 111 23.52 6 1.27 9 Tondihal 1270 93 7.32 33 2.60 10 Mallapur 0 0 0.00 0 0.00 11 Halkawatgi 1718 483 28.11 19 1.11 12 Palgal Dinni 578 161 27.85 30 5.19 13 Tumbalgaddi 423 58 13.71 16 3.78 14 Rampur 531 131 24.67 0 0.00 15 Nagarhal 3880 904 23.30 182 4.69 16 Bhogapur 773 281 36.35 6 0.78 17 Baiyapur 1331 504 37.87 16 1.20 18 Khairwadgi 2044 655 32.05 225 11.01 19 Bandisunkapur 479 9 1.88 16 3.34 20 Bommanhal 1108 221 19.95 4 0.36 21 Sajjalagudda 1100 73 6.64 436 39.64 22 Komnur 779 79 10.14 111 14.25 23 Lukkihal(Big) 646 339 52.48 0 0.00 24 Lukkihal(Small) 921 182 19.76 5 0.54 25 Uppar Nandihal 1151 94 8.17 58 5.04 26 Killar Hatti 1413 490 34.68 0 0.00 27 Ashihal 2162 1775 82.10 150 6.94 28 Advibhavi (Mudgal) 1531 130 8.49 253 16.53 29 Kannapur Hatti 2250 791 35.16 117 5.20 30 Mudgal(Rural) 2235 1271 56.87 21 0.94 31 Jantapur 1150 262 22.78 0 0.00 32 Yerdihal(Khurd) 703 76 10.81 29 4.13 33 Yerdihal(Big) 887 355 40.02 54 6.09 34 Amdihal 2736 886 32.38 10 0.37 35 Bellihal 476 38 7.98 34 7.14 36 Kansavi 1778 395 22.22 83 4.67 37 Adapur 1022 228 22.31 126 12.33 38 Komlapur 951 59 6.20 79 8.31 39 Ramatnal 853 81 9.50 55 -
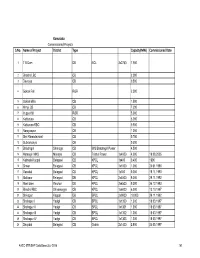
Karnataka Commissioned Projects S.No. Name of Project District Type Capacity(MW) Commissioned Date
Karnataka Commissioned Projects S.No. Name of Project District Type Capacity(MW) Commissioned Date 1 T B Dam DB NCL 3x2750 7.950 2 Bhadra LBC CB 2.000 3 Devraya CB 0.500 4 Gokak Fall ROR 2.500 5 Gokak Mills CB 1.500 6 Himpi CB CB 7.200 7 Iruppu fall ROR 5.000 8 Kattepura CB 5.000 9 Kattepura RBC CB 0.500 10 Narayanpur CB 1.200 11 Shri Ramadevaral CB 0.750 12 Subramanya CB 0.500 13 Bhadragiri Shimoga CB M/S Bhadragiri Power 4.500 14 Hemagiri MHS Mandya CB Trishul Power 1x4000 4.000 19.08.2005 15 Kalmala-Koppal Belagavi CB KPCL 1x400 0.400 1990 16 Sirwar Belagavi CB KPCL 1x1000 1.000 24.01.1990 17 Ganekal Belagavi CB KPCL 1x350 0.350 19.11.1993 18 Mallapur Belagavi DB KPCL 2x4500 9.000 29.11.1992 19 Mani dam Raichur DB KPCL 2x4500 9.000 24.12.1993 20 Bhadra RBC Shivamogga CB KPCL 1x6000 6.000 13.10.1997 21 Shivapur Koppal DB BPCL 2x9000 18.000 29.11.1992 22 Shahapur I Yadgir CB BPCL 1x1300 1.300 18.03.1997 23 Shahapur II Yadgir CB BPCL 1x1301 1.300 18.03.1997 24 Shahapur III Yadgir CB BPCL 1x1302 1.300 18.03.1997 25 Shahapur IV Yadgir CB BPCL 1x1303 1.300 18.03.1997 26 Dhupdal Belagavi CB Gokak 2x1400 2.800 04.05.1997 AHEC-IITR/SHP Data Base/July 2016 141 S.No. Name of Project District Type Capacity(MW) Commissioned Date 27 Anwari Shivamogga CB Dandeli Steel 2x750 1.500 04.05.1997 28 Chunchankatte Mysore ROR Graphite India 2x9000 18.000 13.10.1997 Karnataka State 29 Elaneer ROR Council for Science and 1x200 0.200 01.01.2005 Technology 30 Attihalla Mandya CB Yuken 1x350 0.350 03.07.1998 31 Shiva Mandya CB Cauvery 1x3000 3.000 10.09.1998 -

Modelling of Short Duration Isopluvial Map for Raichur District Karnataka
IJSART - Volume 5 Issue 4 –APRIL 2019 ISSN [ONLINE]: 2395-1052 Modelling of Short Duration Isopluvial Map For Raichur District Karnataka Mohammed Badiuddin Parvez1, M .Inayathulla2 1Dept of Civil Engineering 2Professor, Dept of Civil Engineering 1, 2 UVCE, Bangalore University, Bangalore ,Karnataka, India. Abstract- Everyoneacknowledges that it rains, runoff is The scope of this study was to predict rainfall generated for a design point of view we should know how intensity for the stations using the data of 1998 to 2016 spread much and how often it rains on our project in Raichur District by using Log Normal distribution and location.Estimation of rainfall intensity is commonly required Develop Isopluvial Maps of different duration and return for the design of hydraulic and water resources engineering period. control structures. The present study aimed the Estimation of rainfall intensityin Raichur District using twenty five Rain II. MATERIALS AND METHODS gauge Station with 19 years of rainfall data (1998 to 2016). Log Normal Distribution, techniques are used to derived the 2.1 Study Area rainfall intensity values of 2,5,10,15,30,60,120,720,1440 minutes of rainfall duration with different return period. The short duration IDF using daily rainfall data are presented, which is input for water resources projects. Isopluvial maps were developed for 25years, 50years, 75years and 100years return period Keywords- Isopluvial Maps, Log Normal Distribution, Rainfall Duration, Return Period, Rainfall Intensity. I. INTRODUCTION Short-term, high-intensity rainfall that occurs in inland areas with poor drainage often produces urban flash floods. Densely populated areas have a high risk for flash floods. -
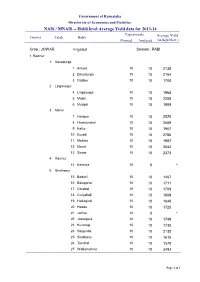
NAIS / MNAIS -- Hobli Level Average Yield Data for 2013-14 Experiments Average Yield District Taluk Hobli Planned Analysed (In Kgs/Hect.)
Government of Karnataka Directorate of Economics and Statistics NAIS / MNAIS -- Hobli level Average Yield data for 2013-14 Experiments Average Yield District Taluk Hobli Planned Analysed (in Kgs/Hect.) Crop : JOWAR Irrigated Season : RABI 1 Raichur 1 Devadurga 1 Arkera 10 10 2135 2 Devadurga 10 10 2164 3 Gabbur 10 10 1708 2 Lingasugur 4 Lingasugur 10 10 1968 5 Maski 10 10 2208 6 Mudgal 10 10 1909 3 Manvi 7 Halapur 10 10 2575 8 Hirekotnakal 10 10 2659 9 Kallur 10 10 1907 10 Kuradi 10 10 2786 11 Mallata 10 10 1987 12 Manvi 10 10 2042 13 Sirwar 10 10 2373 4 Raichur 14 Kalmala 10 0 * 5 Sindhanur 15 Badarli 10 10 1467 16 Balaganur 10 10 1711 17 Gorebal 10 10 1709 18 Gunjahalli 10 10 1809 19 Hedaginal 10 10 1648 20 Hooda 10 10 1720 21 Jalihal 10 0 * 22 Jawalgera 10 10 1749 23 Kunnatgi 10 10 1735 24 Salgunda 10 10 2132 25 Sindhanur 10 10 1615 26 Turvihal 10 10 1578 27 Walkamdinni 10 10 3492 Page 1 of 3 Experiments Average Yield District Taluk Hobli Planned Analysed (in Kgs/Hect.) Crop : GRAM Irrigated Season : RABI 1 Raichur 1 Lingasugur 28 Lingasugur 10 10 617 29 Mudgal 10 10 556 2 Manvi 30 Halapur 10 10 670 31 Hirekotnakal 10 10 693 32 Kallur 10 10 1113 33 Kuradi 10 10 1265 34 Manvi 10 10 945 35 Sirwar 10 10 1102 3 Raichur 36 Kalmala 10 0 * 4 Sindhanur 37 Badarli 10 10 1281 38 Balaganur 10 10 1103 39 Jawalgera 10 10 1122 40 Salgunda 10 10 1169 41 Sindhanur 10 10 1155 42 Turvihal 10 10 1138 Crop : SUNFLOWER Irrigated Season : RABI 1 Raichur 1 Lingasugur 43 Gurgunta 10 10 973 44 Lingasugur 10 10 973 45 Maski 10 10 1066 46 Mudgal 10 10 1140 2 -

Raichur Rural
MLA Constituency Name Mon Aug 24 2015 Raichur Rural Elected Representative :Thipparaju Political Affiliation :Bharatiya Janata Party Number of Government Schools in Report :197 KARNATAKA LEARNING PARTNERSHIP This report is published by Karnataka Learning Partnership to provide Elected Representatives of Assembly and Parliamentary constituencies information on the state of toilets, drinking water and libraries in Government Primary Schools. e c r s u k o o S t o r e l e B i t o a h t t t T e i e W l l i n i W g o o o y y n T T i r r m k s a a s r r l m y n r i b b i o o r i i District Block Cluster School Name Dise Code C B G L L D RAICHUR MANVI GORKAL GHPS BAIL MARCHED 29060601101 Tap Water RAICHUR MANVI GORKAL GHPS GORKAL 29060604601 Tap Water RAICHUR MANVI GORKAL GHPS RAJALABANDA 29060612701 Tap Water RAICHUR MANVI GORKAL GHPS SUNKESWAR 29060613904 Tap Water RAICHUR MANVI KALLUR G LPS LAXMI NAGAR CAMP KALLUR 29060608410 Tap Water RAICHUR MANVI KURDI GHPS AROLI 29060600701 Tap Water RAICHUR MANVI KURDI GHPS GIRLS KURDI 29060609602 Tap Water RAICHUR MANVI KURDI GHPS KAMBALANETHI 29060608501 Tap Water RAICHUR MANVI KURDI GHPS KURADI 29060609601 Tap Water RAICHUR MANVI KURDI GHPS RAJOLLI 29060612901 Tap Water RAICHUR MANVI KURDI GHPS VALAKAMDINNI 29060615201 Tap Water RAICHUR MANVI KURDI GLPS ADAVIKHANPUR 29060600101 Tap Water RAICHUR MANVI KURDI GLPS KURDI 29060609610 Tap Water RAICHUR MANVI KURDI GLPS URDU AROLI 29060600702 Others RAICHUR MANVI KURDI GLPS URDU KURADI 29060609607 Others MLA Constituency Name => Raichur Rural e c r s u k -

Communication Directory-2017.Pmd
COMMUNICATION DIRECTORY – 2017 Chancellor’s Office (O) 080 - 22254102 / 22254109 Rajbhavan, Bangalore (Fax) 080 - 22258150 Pro Chancellor’s (O) 080 - 22383418 Office Minister of Agriculture (Fax) 080 - 22034783 Vidhan Soudha, GOK, Bangalore BOARD OF MANAGEMENT Dr. P.M. Salimath (O) 08532 - 221444 Vice – Chancellor (R) 08532 - 220153 (Fax) 08532 - 220444 (M) 9480636300 / 9902480022 E-Mail: [email protected] [email protected] Principle Secretary (O) 080 - 22250284 / 22032595 Department of Agriculture (Fax) 080-22251420 Principle Secretary (O) 080 - 22365687 / 23515855 Department of Horticulture Principle / Deputy Secretary Department of Finance (O) 080 - 22282590/ 22252078 Secretary Department of (O) 080 - 22254377 Forest & Ecology (Fax) 080 - 22254377 Dr. M. Shekhargouda (M) 8951529119 Post: Herur, tq: Gangavathi, Email:[email protected] Dist: Koppal -583 227 - 1 - Mrs. Hemavathi Lankesh (M) 9972769517 / 9880205035 Sakin Guldal, Hanuwal Post, tq: Gangavathi, Dist: Koppal Mr. Amaresh Ballidava (M) 9880768873 Bapuji Oni, Deodurga, Raichur-584 111l Mr. Siddappa Bhandari (M) 9880074579 / 7795689567 #29A, Kallur Road Email: [email protected] Kalmala - 584 136 Tq. Dist. Raichur Mr. Veeranagouda (M) 9449822633 Post: Nalavara, Chitapur taluk, Kalaburagi - 585 211 Dr. D.M. Chandargi (O) 08532-220157 Member Secretary &Registrar (Fax) 08532-220440 UAS, Raichur (M) 9480696302 Email : [email protected] - 2 - OFFICE OF THE VICE-CHANCELLOR Ph.No: 08532-221444 Fax No08532-220444 E-mail: [email protected] / [email protected] Dr. P. M. Salimath (M) 9480696300 / 9902480022 Vice-Chancellor (R) 08532 - 220153 (O) 08532 - 221444 E-mail: [email protected] Mr. Suryakanth (M) 9019949701 Secretary (O) 08532 - 221444 E-mail: [email protected] Mr. Shafeed Sullad (M) 9480696339(R) 9448777042 Superintendent (Accts) (O) 08532 - 221444 E-mail: [email protected] Mrs. -

Rural Water Supply 07-08 Action Plan
PANCHAYAT RAJ ENGINEERING DIVISION, RAICHUR PROFORMA FOR ACTION PLAN 2007-08 Present water supply Proposed water supply status Scheme Expendi Sl Estimate Gram Panchayat Village/ Habitation ture as on No cost LPCD Taluk TOTAL District 31-3-2007 Remarks HP HP OWS OWS PWSS PWSS MWSS MWSS MWSS MWSS MWSS MWSS for recharging for Supply Levelof Population 2001 Population 2007 Population Amount required Amount required Amount PWSS R/A PWSS MWSSR/A Single phase Single Single phase Single 1 2 3 4 5 6 7 8 9 1011121314151617181920212223 24 25 26 SPILLOVER FOR 2007-08 (STATE SECTOR) 1 RCR RCR Yapaldinni Naradagadde 30 34 3.00 1.77 - - - - - - - - 1 - - - - 1.23 0.00 1.23 Completed 2 RCR RCR Marchetal Pesaldinni 97 107 2.00 0.00 - - 1 - - 114 - - - - - - 1 2.00 0.00 2.00 C-99 In progress 3 RCR RCR Matmari Heerapur 2575 2884 1.25 0.00 1 1 7 - - 40 - A - - - - - 1.25 0.00 1.25 In progress 4 RCR RCR Gillesugur Duganur 1375 1540 2.00 0.00 - 1 4 - - 26 - - - A - - - 2.00 0.00 2.00 C-99 In progress 5 RCR RCR J.Venkatapur Gonal 952 1066 4.50 0.24 - 1 3 - - 30 - - - A - - - 4.26 0.00 4.26 In progress 6 RCR RCR Sagamkunta Mamadadoddi 1125 1260 4.00 0.00 - 1 4 - - 15 - - - R - - - 4.00 0.00 4.00 S.B In progress 7 RCR RCR Kalmala Kalmala 5797 6493 1.00 0.00 2 1 8 - - 23 - A - - - - - 1.00 0.00 1.00 C-99 Completed 8 RCR RCR Mamdapur Nelhal 2242 2511 0.35 0.00 1 1 1 - 1 31 - A - - - - - 0.35 0.00 0.35 S.B- Completed 9 RCR RCR Idapanur Idapanur 4934 5526 4.00 0.00 2 - 8 - - 21 - R - - - - - 4.00 0.00 4.00 S.B In progress 10 RCR RCR Kamalapur Manjarla 1463 1639 1.00 -

Government of Karnataka Directorate of Economics
GOVERNMENT OF KARNATAKA DIRECTORATE OF ECONOMICS AND STATISTICS 1 RASHTRIYA KRISHI BIMA YOJANA (National Agricultural Insurance Scheme) CROP CUTTING EXPERIMENTS CONDUCTED AND AVERAGE YIELD FOR THE YEAR 2007-08 DISTRICT :RAICHUR SEASON:KHARIF CROP :RICE SOURCE:IRRIGATED --------------------------------------------------------------------------- EXPERIMENTS ASSESSED TALUK HOBLI --------------------- YIELD PLANNED | ANALYSED (Kgs/Hect) --------------------------------------------------------------------------- 1 2 3 4 5 --------------------------------------------------------------------------- 1 Devadurga 1 Arkera 12 12 3585 2 Devadurga 12 12 3537 3 Gabbur 12 12 3533 4 Jalahalli 12 12 3818 2 Lingasugur 5 Gurgunta 12 12 2638 6 Lingasugur 12 12 2559 3 Manvi 7 Halapur 12 12 3351 8 Hirekotnakal 12 12 3369 9 Kallur 12 12 3640 10 Kavithala 12 12 3337 11 Kuradi 12 12 3545 12 Mallata 12 12 3401 13 Manvi 12 12 3170 14 Sirwar 12 12 3541 4 Raichur 15 Chandrabanda 12 12 3529 16 Devasugur 12 12 3657 17 Gillesugur 12 12 3724 18 Kalmala 12 12 3587 19 Raichur 12 12 3574 20 Yeragera 12 12 3804 5 Sindhanur 21 Badarli 12 12 2925 22 Balaganur 12 12 2621 23 Gorebal 12 12 2941 24 Gudadur 12 12 2951 25 Gunjahalli 12 12 3056 26 Hedaginal 12 12 2763 27 Hooda 12 12 2760 28 Jalihal 12 12 2884 29 Jawalgera 12 12 2962 30 Kunnatgi 12 12 2654 31 Salgunda 12 12 2765 32 Sindhanur 12 12 2703 33 Turvihal 12 12 2796 34 Walkamdinni 12 12 2761 --------------------------------------------------------------------------- GOVERNMENT OF KARNATAKA DIRECTORATE OF ECONOMICS AND STATISTICS -
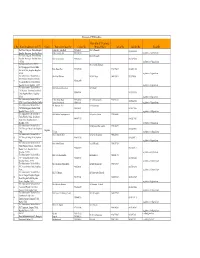
Karnataka ICTC Data Base
Karnataka ICTC Data Base Name of the ICTC Incharge / S. No Name & Address of the ICTC District Name of the Counsellor Contact No Medical Officer Contact No Land line No. Email ID The District Surgeon, District Hospital 1)Shri. R. S. Lingdhalli 9945646629 1 Dr.S,S.Chincolli 08354236100 Bagalkot, Navanagar, Bagalkot District. 2)Shri.Laxman. Jingi 9844038154 [email protected] The District Surgeon, District Hospital Dr.S,S.Chincolli 2 Bagalkot, Navanagar, Bagalkot District. Smt. Savitri Konnur. 9740725218 08354236100 - 587102 [email protected] The Medical Officer I/c PPTCT, Dr.Geetanjali. Katageri Sri, Nijalingappa Medical & HSK 3 Kum: Rajani Rai. 9945297426 9448776044 0830254 1160 Research Cebtre, Bagalkot. Bagalkot - 587102 [email protected] The Administrative Medical Officer, Shri Vittal Dhalwai Dr.M.A.Desai 9448333811 8353220066 Sub Divisional Hospital Jamkhandi, 4 9901421090 Near Ambedkar Circle Jolada Bazar, Bagalkot District, Bagalkot - 587301 [email protected] The Administrative Medical Officer, Smt. Mahanand Shirahatti. Dr.N.Nadaf CHC Rabkavi - Banahatti, Jamkhnadi 5 9880629841 08353230238 Taluk, Bagalkot District, Bagalkot - 587311 [email protected] The Administrative Medical Officer, 1) Shri. Kiran Mardi 9740725565 Dr.A.A.Suryavanshi 9448392122 6 08350280038 KEM General Hospital Mudhol, Mudho 2)Smt.Geeta Kamble 9880891611 [email protected] The Administrative Medical Officer, Sri. Basavaraj Teli. Dr.Vajjaramatti 7 CHC Mahalingapura, Mudhol Taluk, 9880590031 08350273100 Bagalkot District. - 587312 [email protected] The Administrative Medical Officer, Smt.Shobha. Gopalappanavar Dr.Jayashree.Emmi 9902460600 Taluka Hospital Bilagi, Jamakhandi 8 9845967767 08425275421 Road, Bilagi, Bagalkot District. Bagalkot - 587116 [email protected] The Administrative Medical Officer, Dr.Dayanand. -

Ground Water Year Book of Karnataka State 2015-2016
FOR OFFICIAL USE ONLY No. YB-02/2016-17 GROUND WATER YEAR BOOK OF KARNATAKA STATE 2015-2016 CENTRAL GROUND WATER BOARD SOUTH WESTERN REGION BANGALORE NOVEMBER 2016 GROUND WATER YEAR BOOK 2015-16 KARNATAKA C O N T E N T S SL.NO. ITEM PAGE NO. FOREWORD ABSTRACT 1 GENERAL FEATURES 1-10 1.1 Introduction 1.2 Physiography 1.3. Drainage 1.4. Geology RAINFALL DISTRIBUTION IN KARNATAKA STATE-2015 2.1 Pre-Monsoon Season -2015 2 2.2 South-west Monsoon Season - 2015 11-19 2.3 North-east Monsoon Season - 2015 2.4 Annual rainfall 3 GROUND WATER LEVELS IN GOA DURING WATER YEAR 20-31 2015-16 3.1 Depth to Ground Water Levels 3.2 Fluctuations in the ground water levels 4 HYDROCHEMISTRY 32-34 5 CONCLUSIONS 35-36 LIST OF FIGURES Fig. 1.1 Administrative set-up of Karnataka State Fig. 1.2 Agro-climatic Zones of Karnataka State Fig. 1.3 Major River Basins of Karnataka State Fig. 1.4 Geological Map of Karnataka Fig. 2.1 Pre-monsoon (2015) rainfall distribution in Karnataka State Fig. 2.2 South -West monsoon (2015) rainfall distribution in Karnataka State Fig. 2.3 North-East monsoon (2015) rainfall distribution in Karnataka State Fig. 2.4 Annual rainfall (2015) distribution in Karnataka State Fig. 3.1 Depth to Water Table Map of Karnataka, May 2015 Fig. 3.2 Depth to Water Table Map of Karnataka, August 2015 Fig. 3.3 Depth to Water Table Map of Karnataka, November 2015 Fig. 3.4 Depth to Water Table Map of Karnataka, January 2016 Fig. -

Karnataka Circle Cycle III Vide Notification R&E/2-94/GDS ONLINE CYCLE-III/2020 DATED at BENGALURU-560001, the 21-12-2020
Selection list of Gramin Dak Sevak for Karnataka circle Cycle III vide Notification R&E/2-94/GDS ONLINE CYCLE-III/2020 DATED AT BENGALURU-560001, THE 21-12-2020 S.No Division HO Name SO Name BO Name Post Name Cate No Registration Selected Candidate gory of Number with Percentage Post s 1 Bangalore Bangalore ARABIC ARABIC GDS ABPM/ EWS 1 DR1786DA234B73 MONU KUMAR- East GPO COLLEGE COLLEGE Dak Sevak (95)-UR-EWS 2 Bangalore Bangalore ARABIC ARABIC GDS ABPM/ OBC 1 DR3F414F94DC77 MEGHANA M- East GPO COLLEGE COLLEGE Dak Sevak (95.84)-OBC 3 Bangalore Bangalore ARABIC ARABIC GDS ABPM/ ST 1 DR774D4834C4BA HARSHA H M- East GPO COLLEGE COLLEGE Dak Sevak (93.12)-ST 4 Bangalore Bangalore Dr. Dr. GDS ABPM/ ST 1 DR8DDF4C1EB635 PRABHU- (95.84)- East GPO Shivarama Shivarama Dak Sevak ST Karanth Karanth Nagar S.O Nagar S.O 5 Bangalore Bangalore Dr. Dr. GDS ABPM/ UR 2 DR5E174CAFDDE SACHIN ADIVEPPA East GPO Shivarama Shivarama Dak Sevak F HAROGOPPA- Karanth Karanth (94.08)-UR Nagar S.O Nagar S.O 6 Bangalore Bangalore Dr. Dr. GDS ABPM/ UR 2 DR849944F54529 SHANTHKUMAR B- East GPO Shivarama Shivarama Dak Sevak (94.08)-UR Karanth Karanth Nagar S.O Nagar S.O 7 Bangalore Bangalore H.K.P. Road H.K.P. Road GDS ABPM/ SC 1 DR873E54C26615 AJAY- (95)-SC East GPO S.O S.O Dak Sevak 8 Bangalore Bangalore HORAMAVU HORAMAVU GDS ABPM/ SC 1 DR23DCD1262A44 KRISHNA POL- East GPO Dak Sevak (93.92)-SC 9 Bangalore Bangalore Kalyananagar Banaswadi GDS ABPM/ OBC 1 DR58C945D22D77 JAYANTH H S- East GPO S.O S.O Dak Sevak (97.6)-OBC 10 Bangalore Bangalore Kalyananagar Kalyananagar GDS ABPM/ OBC 1 DR83E4F8781D9A MAMATHA S- East GPO S.O S.O Dak Sevak (96.32)-OBC 11 Bangalore Bangalore Kalyananagar Kalyananagar GDS ABPM/ UR 1 DR26EE624216A1 DHANYATA S East GPO S.O S.O Dak Sevak NAYAK- (95.8)-UR 12 Bangalore Bangalore St. -

2007-08 40 (Ii) Explanatory Notes 43 No
GOVERNMENT OF KARNATAKA FINANCE ACCOUNTS 2007-2008 TABLE OF CONTENTS Reference to page Certificate of the Comptroller and Auditor General of India 3 Introductory 4 PART 1 - SUMMARISED STATEMENTS No. 1 - Summary of Transactions 8 - Explanatory Notes 20 No. 2 - Capital Outlay outside the Revenue Account (i) Progressive Capital Outlay to end of 2007-08 40 (ii) Explanatory Notes 43 No. 3 - (i) Financial results of Irrigation Works 46 (ii) Financial results of Electricity Schemes 47 No. 4 - Debt Position - (i) Statement of Borrowings 48 (ii) Other Obligations 50 (iii) Service of Debt 51 No. 5 - Loans and Advances by State Government - (i) Statement of Loans and Advances 52 (ii) Recoveries in arrears 53 No. 6 - Guarantees given by the Government of Karnataka in respect of loans etc., raised by Statutory Corporations, Government Companies, Local Bodies and other Institutions 56 No. 7 - Cash balances and investments of cash balances 62 No. 8 - Summary of balances under Consolidated Fund, Contingency Fund and Public Account 64 PART II - DETAILED ACCOUNTS AND OTHER STATEMENTS SECTION A - REVENUE AND EXPENDITURE No. 9 - Statement of revenue and expenditure for the year 2007-08 expressed as a percentage of total revenue / total expenditure 71 No. 10 - Statement showing the distribution between charged and voted expenditure 74 No. 11 - Detailed account of revenue receipts and capital receipts by minor heads 75 No. 12 - Detailed account of expenditure by minor heads 95 No. 13 - Detailed statement of capital expenditure during and to end of the year 2007-08 132 No. 14 - Details of investments of Government in Statutory Corporations, Government Companies, Other Joint Stock companies, Co-operative Banks and Societies etc., to end of 2007-08 251 No.