Spondence Address: A63L/7088 (2006.01) MUETING, RAASCH & GEBHARDT, P.A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nucleic Acid Nanostructures–DNA and RNA Nanoparticles Lekhana S, Kanishka B, Sneha Thakur*
REVIEW ARTICLE Nucleic Acid Nanostructures–DNA and RNA Nanoparticles Lekhana S, Kanishka B, Sneha Thakur* Department of Pharmacy, Bojjam Narsimhulu Pharmacy College for Women, Saidabad, Hyderabad-500059, Telangana, India Received: 20th March, 2020; Revised: 19th April, 2020; Accepted: 21st May, 2020; Available Online: 25th June, 2020 ABSTRACT Nanotechnology is the field of science that encompasses the production of nanoparticles at nanoatomic scale. The nanoparticles size governs wide range of therapeutic properties. The nanostructures when formulated using deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) materials can result in enhanced hydrophobic properties, shelf life, and also may enhance targeted drug delivery. Structured or dynamic DNA nanostructures involve basic nucleotide pairing with protein backbone. The DNA nanostructures produced by bottom to top approach are currently being applied in medicine, cancer, ophthalmic drug delivery, and also for diagnosis and treatment. They involve complimentary DNA base pairing (AT/GC) for strong interaction resulting in formulation of structures with 10 to 50 mm size. RNA nanostructures are multifunctional manipulative structures synthesized using a bottom-up approach, which results in modulated physiochemical properties and enhanced stability. RNA nanoparticles are currently exploited in the field of cancer. Both DNA and RNA nanostructures are 2D/3D multidimensional models, which are novel structures in the field of nanoscience. DNA/RNA nanostructures can be identified by the Killer Killiani test, quantitative estimation, and also spectroscopic methods like UV, SDS-Page, XRD, SEM, and DLS techniques can be applied. The wide range of applications of these DNA/RNA nanostructures are currently considered as an area of interest in field of therapeutics by most of the researchers. -

Molecular Basis of Angiogenesis and Neuroprotection by Angiogenin By
Molecular Basis of Angiogenesis and Neuroprotection by Angiogenin By Trish T. Hoang A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biochemistry) at the UNIVERSITY OF WISCONSIN–MADISON 2016 Date of final oral examination: May 09, 2016 The dissertation is approved by the following members of the Final Oral Committee: Ronald T. Raines, Henry Lardy Professor of Biochemistry, Biochemistry and Chemistry Jeffrey A. Johnson, Professor, Pharmaceutical Sciences David J. Pagliarini, Associate Professor, Biochemistry Samuel E. Butcher, Professor, Biochemistry Nader Sheibani, Professor, Ophthalmology and Visual Sciences ProQuest Number:10189602 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. ProQuest 10189602 Published by ProQuest LLC ( 2018). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code Microform Edition © ProQuest LLC. ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, MI 48106 - 1346 i Molecular Basis of Angiogenesis and Neuroprotection by Angiogenin Trish Truc Hoang Under the Supervision of Professor Ronald T. Raines at the University of Wisconsin–Madison Cancer and neurodegeneration are disorders with profound impact on human health. Cancer results from uncontrolled cell growth, whereas neurodegeneration is caused by premature neuronal cell death. Although these disease mechanisms seem to be at opposite ends of a spectrum, an increasing number of cellular and molecular studies have linked the two disorders. -

Assembly of Phi29 Prna Nanoparticles for Gene Or Drug
Assembly of Phi29 pRNA Nanoparticles for Gene or Drug Delivery and for Application in Nanotechnology and Nanomedicine A dissertation submitted to The Division of Research and Advanced Studies, University of Cincinnati In partial fulfillment of the requirements for the degree of Doctor of Philosophy In the Biomedical Engineering Program School of Energy, Environmental, Biological and Medical Engineering College of Engineering and Applied Science 2012 by Yi Shu M.S. Chinese Center for Disease Control and Prevention (CCDC), China, 2007 B.S. Lanzhou University, China, 2004 Committee Chair: Jing-Hui Lee, Ph.D Co-chair: Peixuan Guo, Ph.D ABSTRACT RNA nanotechnology is to extract defined RNA structure motifs and tertiary interactions, apply them as the building blocks to self-assemble nano-scaled scaffolds with rational designs, and incorporate functional molecules such as siRNA, ribozyme, aptamer and therapeutical compounds to form functionalized RNA nanoparticles. Bacteriophage phi29 packaging RNA (pRNA) has two defined domains: the 5’/3’-end helical domain and the interlocking loop region which is located at the central part of the pRNA sequence. pRNA dimer is formed by hand-in-hand interaction via 4-bp interlocking base pairing. The dimeric pRNA nanoparticle has been shown to be an efficient vector for the specific delivery of small interfering RNA (siRNA) into specific cancer or viral infected cells. However, there are several problems hindering the therapeutic applications of pRNA nanoparticles. In this thesis, I will try to address: 1) The problem of large-scale synthesis of longer RNA molecules. Industrial scale production of RNA by chemical synthesis is limited to ~ 80nt. -

BIOINF-2006-1117 Revision 3
Bioinformatics Advance Access published January 31, 2007 S.K.L NG and S.K MISHRA large internal loops or bulges especially large asymmetric ones from the hairpins that obviates the use of comparative genomics infor- (Ambros et al., 2003). Apparently, mere application of simple align- mation. The SVM classifier model trained with the experimental do- ment queries and positive-selection rules is likely to overlook novel main knowledge and binary-labeled feature vectors, recovered 71% of families lacking clear homologues to published mature miRNAs. the positive pre-miRs with a remarkably low false-positive rate of ~3%. Advanced comparative approaches like MiRscan (Lim et al., 2003b; It also predicted ~50 to 100 novel pre-miRs for several species; ~30% Lim et al., 2003a), MIRcheck (Jones-Rhoades and Bartel 2004), miR- of these were previously experimentally validated. The validation rate Finder (Bonnet et al., 2004a), miRseeker (Lai et al., 2003), findMiRNA among the predicted cases that were conserved in ≥1 other species was (Adai et al., 2005), PalGrade (Bentwich et al., 2005), and MiRAlign higher at ~60%; many had not been detected by comparative genomics (Wang et al., 2005) have systematically exploit the greater availability approaches. The 3SVM (Xue et al., 2005) improved the performances of sequenced genomes for eliminating the over-represented false- to ~90.00% for human and up to 90.00% in other species. Albeit its positives. Cross-species sequence conservation based on computation- methodological simplicity, promising performances, and independence ally intensive multiple genome alignments is a powerful approach for of comparative genomics information, 3SVM was largely limited to genome-wide screening of phylogentically well conserved pre-miRs classifying RNA sequences that fold into secondary structures without between closely related species. -

Non-Viral Vectors for Gene Therapy
VOLUME EIGHTY NINE ADVANCES IN GENETICS ADVANCES IN GENETICS, VOLUME 89 Serial Editors Theodore Friedmann University of California at San Diego, School of Medicine, USA Jay C. Dunlap The Geisel School of Medicine at Dartmouth, Hanover, NH, USA Stephen F. Goodwin University of Oxford, Oxford, UK VOLUME EIGHTY NINE ADVANCES IN GENETICS Nonviral Vectors for Gene Therapy Physical Methods and Medical Translation Edited by LEAF HUANG Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, University of North Carolina at Chapel Hill, Eshelman School of Pharmacy, Chapel Hill, NC, USA DEXI LIU Department of Pharmaceutical and Biomedical Sciences, University of Georgia College of Pharmacy, Athens, GA, USA ERNST WAGNER Munich Center for System-based Drug Research, Center for Nanoscience, Ludwig-Maximilians-Universitat,€ Munich, Germany AMSTERDAM • BOSTON • HEIDELBERG • LONDON NEW YORK • OXFORD • PARIS • SAN DIEGO SAN FRANCISCO • SINGAPORE • SYDNEY • TOKYO Academic Press is an imprint of Elsevier Academic Press is an imprint of Elsevier 225 Wyman Street, Waltham, MA 02451, USA 525 B Street, Suite 1800, San Diego, CA 92101–4495, USA 125 London Wall, London, EC2Y 5AS, UK The Boulevard, Langford Lane, Kidlington, Oxford OX5 1GB, UK First edition 2015 Copyright © 2015 Elsevier Inc. All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Details on how to seek permission, further information about the Publisher’s permissions policies and our arrangements with organizations such as the Copyright Clearance Center and the Copyright Licensing Agency, can be found at our website: www.elsevier.com/permissions. -

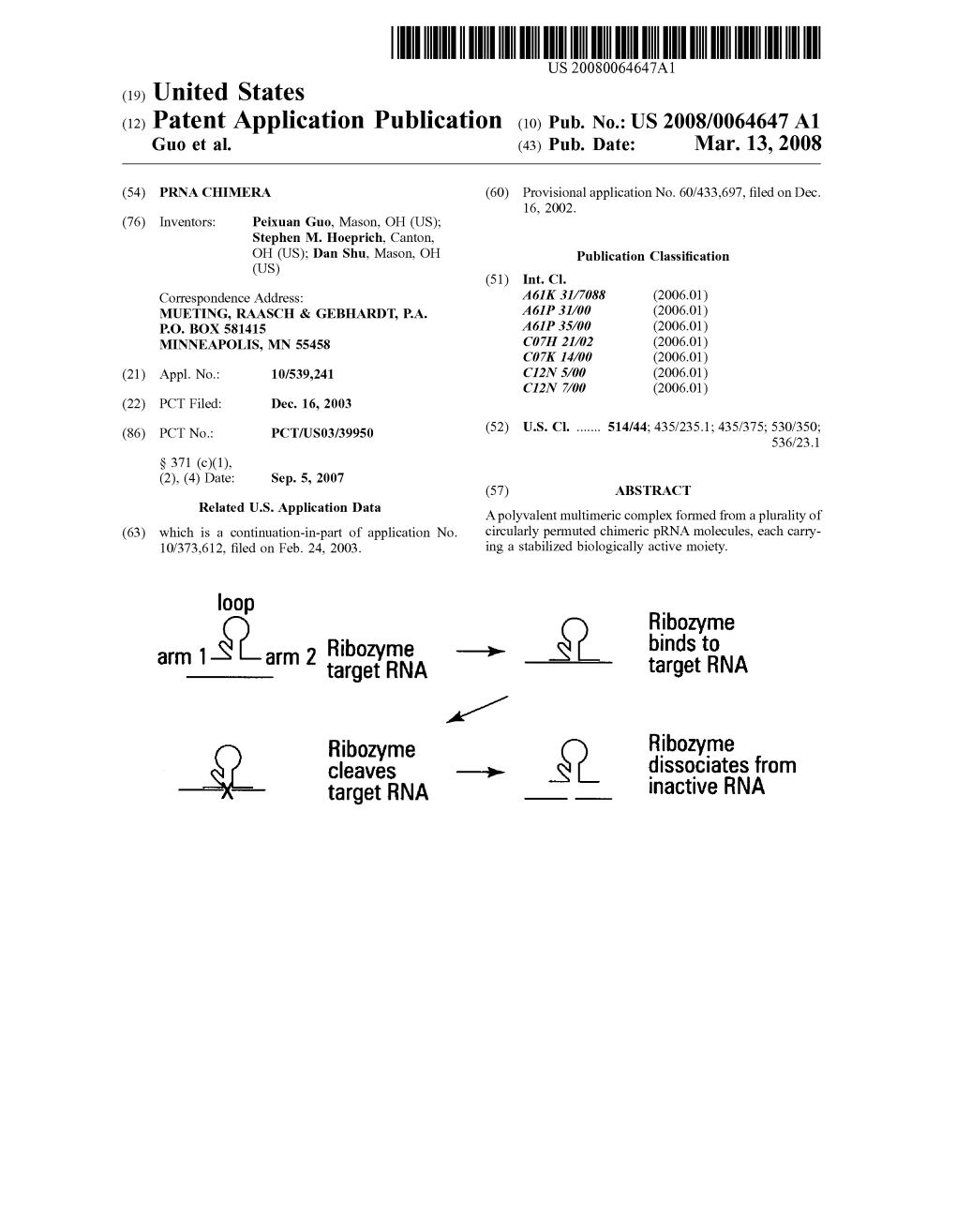
(12) United States Patent (10) Patent No.: US 7.655,787 B2 Guo Et Al
USOO7655787B2 (12) United States Patent (10) Patent No.: US 7.655,787 B2 Guo et al. (45) Date of Patent: Feb. 2, 2010 (54) PRNACHIMERA Zhang et al. Circularly Permuted Viral pRNA Active and Specific in the Packaging of Bateriophage phi29 DNA. Virology 1995, vol. (75) Inventors: Peixuan Guo, West Lafayette, IN (US); 207:442-451. Academic Press Inc. Stephen M. Hoeprich, North Canton, Bailey et al., “Phylogenetic analysis and secondary structure of the OH (US); Dan Shu, West Lafayette, IN Bacillus subtilis bacteriophage RNA required for DNA packaging.” (US) J. Biol. Chem., 1990; 265:22365-70. Betrand et al., “The expression cassette determines the functional (73) Assignee: Purdue Research Foundation, West activity of ribozymes in mammalian cells by controlling their Lafayette, IN (US) intracellular localization. RNA, 1997; 3:75-88. Bramlage et al., “HIV-1 as a target for synthetic ribozyne-mediated (*) Notice: Subject to any disclaimer, the term of this inhibition of gene expression: site selection and inhibition in cell patent is extended or adjusted under 35 culture.” Nucleic Acids Res., 2000; 28(21):4059-4067. U.S.C. 154(b) by 261 days. Cech et al., “Selecting apt RNAs for NMR.” RNA, 1996; 2,625-627. Chen et al., “Magnesium-induced conformational change of packag (21) Appl. No.: 10/373,612 ing RNA for procapsid recognition and binding during phage phi29 DNA encapsidation.” J. Virol., 1997: 71:495-500. (22) Filed: Feb. 24, 2003 Chen et al., “Sequential Action of Six Virus-Encloded DNA-Packag ing RNAs during Phage d29 Genomic DNA Translocation.” J. Virol. (65) Prior Publication Data 1997; 71(5):3864-3871. -

(12) United States Patent (10) Patent No.: US 8,088,912 B2 Guo Et Al
USOO8088912B2 (12) United States Patent (10) Patent No.: US 8,088,912 B2 Guo et al. (45) Date of Patent: *Jan. 3, 2012 (54) PRNACHIMERA WO WO O2, 16596 A2 2?2002 WO WO O2, 16596 A3 2, 2002 WO WO 2005/OO3293 A2 1, 2005 (75) Inventors: Peixuan Guo, Mason, OH (US); WO WO 2005/OO3293 A3 1, 2005 Stephen M. Hoeprich, Canton, OH WO WO 2005/035760 A2 4, 2005 (US); Dan Shu, Mason, OH (US) WO WO 2005/035760 A3 4, 2005 WO WO 2007/O16507 A2 2, 2007 (73) Assignee: Purdue Research Foundation, West WO WO 2007/O16507 A3 2/2007 Lafayette, IN (US) OTHER PUBLICATIONS (*) Notice: Subject to any disclaimer, the term of this Aggarwal et al., “Biodegradable Alginate Microspheres as a Delivery patent is extended or adjusted under 35 System for Naked DNA.” Can. J. Vet. Res, 1999; 63:148-152. U.S.C. 154(b) by 1086 days. Bailey et al., “Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging.” This patent is Subject to a terminal dis J. Biol. Chem., 1990; 265:22365-70. claimer. Bazinetet al., “The DNA translocating vertex of dsDNA bacterioph age.” Ann. Rev. Microbiol., 1985:39:109-129. (21) Appl. No.: 10/539,241 Becerril et al., “Toward selection of internalizing antibodies from phage libraries.” Biochem. Biophy's. Res. Commun., 1999; 255:386 (22) PCT Filed: Dec. 16, 2003 393. Bergelson et al., “Isolation of a common receptor for Coxsackie B (86). PCT No.: PCT/USO3/399.50 viruses and adenoviruses 2 and 5.” Science, 1997:275(5304): 1320 1323. -

Bacteriophages As Therapeutic and Diagnostic Vehicles in Cancer
pharmaceuticals Review Bacteriophages as Therapeutic and Diagnostic Vehicles in Cancer Valentina Foglizzo 1,2 and Serena Marchiò 1,2,* 1 Department of Oncology, University of Torino, 10060 Candiolo, Italy; [email protected] 2 Candiolo Cancer Institute, FPO-IRCCS, 10060 Candiolo, Italy * Correspondence: [email protected] Abstract: Evolution of nanomedicine is the re-design of synthetic and biological carriers to implement novel theranostic platforms. In recent years, bacteriophage research favors this process, which has opened up new roads in drug and gene delivery studies. By displaying antibodies, peptides, or proteins on the surface of different bacteriophages through the phage display technique, it is now possible to unravel specific molecular determinants of both cancer cells and tumor-associated mi- croenvironmental molecules. Downstream applications are manifold, with peptides being employed most of the times to functionalize drug carriers and improve their therapeutic index. Bacteriophages themselves were proven, in this scenario, to be good carriers for imaging molecules and therapeutics as well. Moreover, manipulation of their genetic material to stably vehiculate suicide genes within cancer cells substantially changed perspectives in gene therapy. In this review, we provide exam- ples of how amenable phages can be used as anticancer agents, especially because their systemic administration is possible. We also provide some insights into how their immunogenic profile can be modulated and exploited in immuno-oncology for vaccine production. Keywords: biopanning; drug delivery; gene therapy; phage display; targeting Citation: Foglizzo, V.; Marchiò, S. Bacteriophages as Therapeutic and Diagnostic Vehicles in Cancer. 1. Introduction Pharmaceuticals 2021, 14, 161. Cancer is a plague worldwide, affecting around 9.6 million people annually and https://doi.org/10.3390/ph14020161 accounting for 1 in every 6 deaths, globally. -

Recent Advances in Mirna Delivery Systems
University of Massachusetts Medical School eScholarship@UMMS Open Access Publications by UMMS Authors 2021-01-20 Recent Advances in miRNA Delivery Systems Ishani Dasgupta University of Massachusetts Medical School Et al. Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/oapubs Part of the Molecular Biology Commons, Nucleic Acids, Nucleotides, and Nucleosides Commons, and the Therapeutics Commons Repository Citation Dasgupta I, Chatterjee A. (2021). Recent Advances in miRNA Delivery Systems. Open Access Publications by UMMS Authors. https://doi.org/10.3390/mps4010010. Retrieved from https://escholarship.umassmed.edu/oapubs/4554 Creative Commons License This work is licensed under a Creative Commons Attribution 4.0 License. This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in Open Access Publications by UMMS Authors by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. Perspective Recent Advances in miRNA Delivery Systems Ishani Dasgupta 1,† and Anushila Chatterjee 2,*,† 1 Horae Gene Therapy Center, Department of Pediatrics, University of Massachusetts Medical School, Worcester, MA 01605, USA; [email protected] 2 Department of Immunology and Microbiology, University of Colorado School of Medicine, Aurora, CO 80045, USA * Correspondence: [email protected] † These authors contributed equally to this work. Abstract: MicroRNAs (miRNAs) represent a family of short non-coding regulatory RNA molecules that are produced in a tissue and time-specific manner to orchestrate gene expression post-transcription. MiRNAs hybridize to target mRNA(s) to induce translation repression or mRNA degradation.