Investigation of Genetic Variation Among Turkish Populations of Andricus Lignicola Using Mitochondrial Cytochrome B Gene Sequence Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
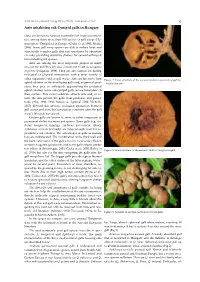
Ants Inhabiting Oak Cynipid Galls in Hungary
North-Western Journal of Zoology 2020, vol.16 (1) - Correspondence: Notes 95 Ants inhabiting oak Cynipid galls in Hungary Oaks are known to harbour extremely rich insect communi- ties, among them more than 100 species of gall wasps (Hy- menoptera: Cynipidae) in Europe (Csóka et al. 2005, Melika 2006). Some gall wasp species are able to induce large and structurally complex galls that can sometimes be abundant on oaks, providing attractive shelters for several arthropod taxa including ant species. Ants are among the most important players in many ecosystems and they are also considered to act as ecosystem engineers (Folgarait, 1998). They are also famous for having ecological or physical interactions with a great variety of other organisms, such as gall wasps. Ants are known to tend Figure 1. Inner structure of the asexual Andricus quercustozae gall in- aphid colonies on the developing galls and, as general pred- habited by ants. ators, they prey on arthropods approaching the protected aphid colonies. Some oak cynipid galls secrete honeydew on their surface. This sweet substrate attracts ants and, in re- turn, the ants protect the galls from predators and parasi- toids (Abe, 1988, 1992; Inouye & Agrawal 2004; Nicholls, 2017). Beyond this obvious ecological interaction between gall wasps and ants, this association continues after the gall wasp’s life cycle has ceased. Certain galls are known to serve as either temporary or permanent shelter for many ant species. Some galls (e.g. An- dricus hungaricus (Hartig), Andricus quercustozae (Bosc), Aphelonyx cerricola (Giraud)) are large enough even for re- productive ant colonies. The advantages of galls as nesting logs are multifaceted. -

The Population Biology of Oak Gall Wasps (Hymenoptera:Cynipidae)
5 Nov 2001 10:11 AR AR147-21.tex AR147-21.SGM ARv2(2001/05/10) P1: GSR Annu. Rev. Entomol. 2002. 47:633–68 Copyright c 2002 by Annual Reviews. All rights reserved THE POPULATION BIOLOGY OF OAK GALL WASPS (HYMENOPTERA:CYNIPIDAE) Graham N. Stone,1 Karsten Schonrogge,¨ 2 Rachel J. Atkinson,3 David Bellido,4 and Juli Pujade-Villar4 1Institute of Cell, Animal, and Population Biology, University of Edinburgh, The King’s Buildings, West Mains Road, Edinburgh EH9 3JT, United Kingdom; e-mail: [email protected] 2Center of Ecology and Hydrology, CEH Dorset, Winfrith Technology Center, Winfrith Newburgh, Dorchester, Dorset DT2 8ZD, United Kingdom; e-mail: [email protected] 3Center for Conservation Science, Department of Biology, University of Stirling, Stirling FK9 4LA, United Kingdom; e-mail: [email protected] 4Departamento de Biologia Animal, Facultat de Biologia, Universitat de Barcelona, Avenida Diagonal 645, 08028 Barcelona, Spain; e-mail: [email protected] Key Words cyclical parthenogenesis, host alternation, food web, parasitoid, population dynamics ■ Abstract Oak gall wasps (Hymenoptera: Cynipidae, Cynipini) are characterized by possession of complex cyclically parthenogenetic life cycles and the ability to induce a wide diversity of highly complex species- and generation-specific galls on oaks and other Fagaceae. The galls support species-rich, closed communities of inquilines and parasitoids that have become a model system in community ecology. We review recent advances in the ecology of oak cynipids, with particular emphasis on life cycle characteristics and the dynamics of the interactions between host plants, gall wasps, and natural enemies. We assess the importance of gall traits in structuring oak cynipid communities and summarize the evidence for bottom-up and top-down effects across trophic levels. -

Contents Hawthorn Dene, 1, 5-Jul-1924
Northern Naturalists’ Union Field Meeting Reports- 1924-2005 Contents Hawthorn Dene, 1, 5-jul-1924 .............................. 10 Billingham Marsh, 2, 13-jun-1925 ......................... 13 Sweethope Lough, 3, 11-jul-1925 ........................ 18 The Sneap, 4, 12-jun-1926 ................................... 24 Great Ayton, 5, 18-jun-1927 ................................. 28 Gibside, 6, 23-jul-1927 ......................................... 28 Langdon Beck, 7, 9-jun-1928 ............................... 29 Hawthorn Dene, 8, 5-jul-1928 .............................. 33 Frosterley, 9 ......................................................... 38 The Sneap, 10, 1-jun-1929 ................................... 38 Allenheads, 11, 6-july-1929 .................................. 43 Dryderdale, 12, 14-jun-1930 ................................. 46 Blanchland, 13, 12-jul-1930 .................................. 49 Devil's Water, 14, 15-jun-1931 ............................. 52 Egglestone, 15, 11-jul-1931 ................................. 53 Windlestone Park, 16, June? ............................... 55 Edmondbyers, 17, 16-jul-1932 ............................. 57 Stanhope and Frosterley, 18, 5-jun-1932 ............. 58 The Sneap, 19, 15-jul-1933 .................................. 61 Pigdon Banks, 20, 1-jun-1934 .............................. 62 Greatham Marsh, 21, 21-jul-1934 ........................ 64 Blanchland, 22, 15-jun-1935 ................................ 66 Dryderdale, 23, ..................................................... 68 Raby Park, -

Torymus Sinensis Against the Chestnut Gall Wasp Dryocosmus Kuriphilus in the Canton Ticino, Switzerland
| January 2011 Evaluating the use of Torymus sinensis against the chestnut gall wasp Dryocosmus kuriphilus in the Canton Ticino, Switzerland Authors Aebi Alexandre, Agroscope ART Schoenenberger Nicola, Tulum SA and Bigler Franz, Agroscope ART Torymus sinensis against the chestnut gall wasp Dryocosmus kuriphilus | January 2011 1 Zürich/Caslano, January 2011 Authors’ affiliation: Alexandre Aebi and Franz Bigler Nicola Schoenenberger Agroscope Reckenholz-Tänikon TULUM SA Research Station ART Via Rompada 40 Biosafety 6987 Caslano Reckenholzstrasse 191 Switzerland 8046 Zürich Tel: +41 91 606 6373 Switzerland Fax: +41 44 606 6376 Tel: +41 44 377 7669 [email protected] Fax: +41 44 377 7201 [email protected] This work was financed by the Swiss Federal Office for the Environment (FOEN) This work was done in collaboration with B. Bellosi and E. Schaltegger (TULUM SA) Cover figure: Empty chestnut gall in Stabio, February 2010 (Picture:TULUM SA) All maps used in figures and appendices (except Fig. 6): ©swisstopo, license number: DV053809.1 Map in figure 6: © Istituto Geografico, De Agostini 1982–1988 ISBN 978-3-905733-20-4 © 2010 ART 2 Torymus sinensis against the chestnut gall wasp Dryocosmus kuriphilus | January 2011 Table of contents Table of contents Abstract 5 1. Introduction 6 2. Mission and methods 7 3. Presence and degree of infestation of Dryocosmus kuriphilus in Switzerland 9 4. Invasion corridors of Dryocosmus kuriphilus towards Switzerland 11 5. Potential economic and ecological damage caused by Dryocosmus kuriphilus in Switzerland 14 6. Release of the parasitoid Torymus sinensis in the Piedmont Region, Italy 17 7. Potential benefits and damage due to the release of Torymus sinensis 18 8. -

The Entomologist's Record and Journal of Variation
. JVASV^iX ^ N^ {/) lSNrNVIN0SHilWS*^S3ldVaan^LIBRARIES SMITHSONIAN INSTITUTION Ni <n - M ^^ <n 5 CO Z ^ ^ 2 ^—^ _j 2 -I RIES SMITHSONIAN INSTITUTION NOIinillSNI NVINOSHilWS S3iyVdan U r- ^ ^ 2 CD 4 A'^iitfwN r: > — w ? _ ISNI NVINOSHilWS SBiyVdan LIBRARIES'SMITHSONIAN INSTITUTION f^ <rt .... CO 2 2 2 s;- W to 2 C/J • 2 CO *^ 2 RIES SMITHSONIAN_INSTITUTlON NOIiniliSNI_NVINOSHilWS S3liiVyan_L; iiSNi"^NViNOSHiiNS S3iyvaan libraries smithsonian'^institution i^ 33 . z I/' ^ ^ (^ RIES SMITHSONIAN INSTITUTION NOIiniliSNI NVINOSHilWS S3lbVHan Li CO — -- — "> — IISNI NVINOSHimS S3IMVHan LIBRARIES SMITHSONIAN INSTITUTION N' 2 -J 2 _j 2 RIES SMITHSONIAN INSTITUTION NOIifllliSNI NVINOSHIIWS SSIMVyail L! MOTITI IT I f\t _NviN0SHiiws'^S3iMvaan libraries'^smithsonian^institution NOlin z \ '^ ^—s^ 5 <^ ^ ^ ^ '^ - /^w\ ^ /^^\ - ^^ ^ /^rf^\ - /^ o ^^^ — x.ii:i2Ji^ o ??'^ — \ii Z ^^^^^""-^ o ^^^^^ -» 2 _J Z -J , ; SMITHSONIAN INSTITUTION NOIXniliSNI NVINOSHillMS $3 I M VH 8 !!_ LI BR = C/> ± O) ^. ? CO I NVINOSHimS S3iaVHan libraries SMITHSONIAN INSTITUTION NOIlf CO ..-. CO 2 Z z . o .3 :/.^ C/)o Z u. ^^^ i to Z CO • z to * z > SMITHS0NIAN_1NSTITUTI0N NOIiniliSNI_NVINOSHimS S3 I d ViJ 8 n_LI B R UJ i"'NViNOSHiiws S3ibvyan libraries smithsonian"^institution Noiir r~ > z r- Z r- 2: . CO . ^ ^ ^ ^ ; SMITHSONIAN INSTITUTION NOIiniliSNI NVINOSHillNS SSiyVMail LI BR CO . •» Z r, <^ 2 z 5 ^^4ii?^^ ^' X^W o ^"^- x life ^<ji; o ^'f;0: i >^ _NVIN0SHiIlMs'^S3iyVdan^LIBRARIEs'^SMITHS0NlAN INSTITUTION NOlif Z \ ^'^ ^-rr-^ 5 CO n CO CO o z > SMITHSONIAN INSTITUTION NOIiniliSNI NVINOSHimS S3 I ^Vd 8 11 LI BR >" _ . z 3 ENTOMOLOGIST'S RECORD AND Journal of Variation Edited by P.A. SOKOLOFF fre s Assistant Editors J.A. -

Interaktionen Zwischen Wirten Und Parasitoiden
Interaktionen zwischen Wirten und Parasitoiden: Nahrungsnetzstruktur, Wirtsspektren und Wirtsfindung am Beispiel der Arten aus Vogelnestern (Insecta: Diptera: Cyclorrhapha und Hymenoptera: Chalcidoidea) Dissertation zur Erlangung des Doktorgrades des Departments Biologie der Fakultät für Mathematik, Informatik und Naturwissenschaften an der Universität Hamburg vorgelegt von Ralph Peters aus Hamburg Dezember 2006 Inhalt Interaktionen zwischen Wirten und Parasitoiden: Nahrungsnetzstruktur, Wirtsspektren und Wirtsfindung am Beispiel der Arten aus Vogelnestern (Insecta: Diptera: Cyclorrhapha und Hymenoptera: Chalcidoidea) Inhalt: 1. Einleitung .......................................................................................................................... 1 2. Material und Methoden .................................................................................................... 4 2.1 Datenerhebung für das Nahrungsnetz / „parasitoid web“ ...................................... 4 2.1.1 Materialsammlung und -weiterbehandlung .................................................................... 4 2.1.2 Weitere Datenerhebung für das „parasitoid web“ ................................................................. 5 2.2 Material und Methoden der Untersuchungen zur Parasitoidenbiologie ................ 6 2.2.1 Morphologische Untersuchungen in der Gattung Dibrachys ................................................ 6 2.2.2 Labornachzuchten zur Erstellung der Wirtsspektren .......................................................... 10 2.2.3 -

Zoologische Mededelingen
ZOOLOGISCHE MEDEDELINGEN UITGEGEVEN DOOR HET RIJKSMUSEUM VAN NATUURLIJKE HISTORIE TE LEIDEN (MINISTERIE VAN CULTUUR, RECREATIE EN MAATSCHAPPELIJK WERK) Deel 53 no. 28 29 juni 1979 A CHARACTER ANALYSIS OF THE SPECIES OF SYNERGUS HARTIG, SECTION II (MAYR, 1872) (HYMENOPTERA, CYNIPIDAE) by A. A. WIEBES - RIJKS Afdeling Systematische Dierkunde der Rijksuniversiteit, Leiden With 83 text-figures ABSTRACT A survey is given of the characters used for the differentiation of the species of Synergus Hartig, classified with section II of Mayr. Special attention is given to the phenology of the species, for each of which the life-cycle is schematized, with differentia- tion in spring- and summer-generation. Two forms, provisionally indicated A and B, could not satisfactorily be identified with any of the known species. Sp. В is a common inquiline of oak-apples; biological observations were made on the larvae. Synergus mutabilis Deitmer, 1924, is synonymized with Synergus albipes Hartig, 1841. The identification of the species of Synergus Hartig, 1840, is notoriously difficult, particularly of those classified in section II (Mayr, 1872). The species of section I, single brooded, that winter in galls as late-stage larvae or pupae and emerge in early summer, are easier to be identified (Eady, 1952). Some of the difficulties with section II may stem from the fact that many species produce two broods in one year, often dissimilar in appearance (Ross, 1951). It should be stated that in all instances the correlations of spring and summer generations were deduced from circumstantial evidence: no specimens were actually reared from one generation to another. Even more confusing than the alternation of broods may be the great variation pre sumably caused by differences in quantity or quality of larval food. -

Index to Cecidology up to Vol. 31 (2016)
Index to Cecidology Up to Vol. 31 (2016) This index has been based on the contents of the papers rather than on their actual titles in order to facilitate the finding of papers on particular subjects. The figures following each entry are the year of publication, the volume and, in brackets, the number of the relevant issue. Aberbargoed Grasslands: report of 2011 field meeting 2012 27 (1) Aberrant Plantains 99 14(2) Acacia species galled by Fungi in India 2014 29(2) Acer gall mites (with illustrations) 2013 28(1) Acer galls: felt galls re-visited 2005 20(2) Acer saccharinum – possibly galled by Dasineura aceris new to Britain 2017 32(1) Acer seed midge 2009 24(1) Aceria anceps new to Ireland 2005 20 (1) Aceria geranii from North Wales 1999 14(2) Aceria heteronyx galling twigs of Norway Maple 2014 29(1) Aceria ilicis (gall mite) galling holm oak flowers in Brittany 1997 12(1) In Ireland 2010 25(1) Aceria mites on sycamore 2005 20(2) Aceria populi galling aspen in Scotland 2000 15(2) Aceria pterocaryae new to the British mite fauna 2008 23(2) Aceria rhodiolae galling roseroot 2013 28(1): 2016 31(1) Aceria rhodiolae in West Sutherland 2014 29(1) Aceria tristriata on Walnut 2007 22(2) Acericecis campestre sp. nov. on Field Maple 2004 19(2) Achillea ptarmica (sneezewort) galled by Macrosiphoniella millefolii 1993 8(2) Acorn galls on red oak 2014 29(1) Acorn stalks: peculiar elongation 2002 17(2) Aculops fuchsiae – a fuchsia-galling mite new to Britain 2008 23 (1) Aculus magnirostris new to Ireland 2005 20 (1) Acumyia acericola – the Acer seed -

Vol. 4 2008 Impressum
Autonome Provinz Bozen - Südtirol Provincia Autonoma di Bolzano - Alto Adige Abteilung Forstwirtschaft – Ripartizione Foreste forest observer vol. 4 2008 Impressum Herausgeber Editore Autonome Provinz Bozen - Südtirol Provincia Autonoma di Bolzano - Alto Adige Abteilung Forstwirtschaft Ripartizione Foreste I-39100 Bozen • Brennerstr. 6 I-39100 Bolzano • Via Brennero 6 Tel. 0471/415300 - 415301 Tel. 0471/415300 - 415301 Fax 0471/415313 Fax 0471/415313 E-Mail [email protected] E-Mail [email protected] Editor in chief Editor in chief Paul Profanter Paul Profanter Direktor Abteilung Forstwirtschaft Direttore Ripartizione Foreste I-39100 Bozen • Brennerstr. 6 I-39100 Bolzano • Via Brennero 6 Redaktion: Redazione: Managing Director Managing Director Florian Blaas Florian Blaas Direktor Amt für Forstverwaltung Direttore Ufficio amministrazione forestale I-39100 Bozen • Brennerstr. 6 I-39100 Bolzano • Via Brennero 6 Koordinator Coordinatore Stefano Minerbi Stefano Minerbi Amt für Forstverwaltung Ufficio amministrazione forestale I-39100 Bozen • Brennerstr. 6 I-39100 Bolzano • Via Brennero 6 Beratung Consulenza Cristina Salvadori Cristina Salvadori Istituto Agrario di S. Michele a /A, Istituto Agrario di S. Michele a /A, Unità operativa – Foreste, Unità operativa – Foreste, I-38010 S. Michele • Via E. Mach 2 I-38010 S. Michele • Via E. Mach 2 Schriftleiter Redattore incaricato Klaus Hellrigl Klaus Hellrigl I-39042 Brixen • Wolkensteinstraße 83 I-39042 Bressanone • Via Wolkenstein 83 Layout & Grafik Layout & Grafica Helga Veleba Helga Veleba Brixen/Wien Bressanone/Vienna Druck Stampa KARO-Druck KARO-Druck Eppan Appiano Copyright: © Landesforstdienst / Servizio Forestale provinciale Abteilung 32 Forstwirtschaft / Ripartizione 32 Foreste Cover picture: Erbsengallen (“Smooth Pea-galls”) der “Weinrosen-Gallwespe” Diplolepis eglanteriae (Hartig 1840) Südtirol: Aicha-Spinges, 19.06.2008, an Blättern von Wildrose Rosa sp. -

Pidb) and Gall Insects
CONTENTS Competition for Young Naturalists ........................................................................................1 Distribution Records ...............................................................................................................1 A Simple Database for Analyzing Oak Gall Wasp Distribution . P.R. Shirley................... 2 BBC Wildlife — Oak Gall Survey .........................................................................................5 Field Meeting — Monks Wood, July 1986 Paul T. Harding & John Pearson ...........................................................................5 Phytophagous Insects Data Bank and Gall Insects Lena K Ward & D.F. Spalding ................................................................................7 E. W. Swanton, O.B.E., A.L.S. — His Notes on Neuroterus ostreus Margaret Hutchinson ...............................................................................................8 Plant Galls from the Isles of Scilly J.P. Bowdrey & B.M. Spooner ..............................................................................10 A List of Insects likely to emerge from Cynipid Galls ..................................P.R. Shirley 13 Aphid Galls and Ladybirds — Peter Cooke ........................................................................20 Plant Galls in Northants — D. Payne ..................................................................................21 Records of Wiltshire Galls — S.A. Manning .......................................................................23 -

Plant Galls on Shotover
I swear (and else may insects prick | Each the female then lays leaf) into a gall, | This girl, for whom your Galls on Shotover eggs on the underside of oak leaves which cause heart is sick | Is three times worth them all. Anywhere the host plant is found can be a worthwhile place the formation of small Tennyson ‘The Talking Oak’ to look for galls, and a walk on Shotover is an ideal way to disc-like Common What are plant galls? begin your search. The woodland edges, hedgerows and spangle galls (♀♀), in late summer and grasslands at Shotover are all worth investigating. Spangle gall If you have ever looked at a tree and been puzzled to see autumn. that it seems to be producing more than one kind of fruit at Galls formed by invertebrates the same time, you have probably seen a plant gall. Silk-button gall (♀♀) is a small ’disc’ with a thickened rim Defined as ‘an abnormal growth produced by a plant or Many invertebrates can cause gall formation. These include and central pit, covered with yellow hairs on the underside other host under the influence of another organism’ (British aphids, mites, psyllids, gall-midges (Cecidomyiidae), gall-flies of the leaves in late summer and autumn. It is caused by the Plant Galls), galls are the host plant’s reaction to some (Tephritidae), sawflies (Symphyta) and especially gall-wasps gall wasp Neuroterus numismalis. kind of infestation, often caused by insects but also by (Cynipidae), as well as a wide range of other invertebrates. fungi (especially rusts and smuts) as well as viruses or Cola nut gall (♀♀). -

Gallwasp Species in Britain
Article (refereed) - postprint Schonrogge, Karsten; Begg, Tracey; Williams, Robin; Melika, George; Randle, Zoe; Stone, Graham N.. 2012 Range expansion and enemy recruitment by eight alien gall wasp species in Britain. Insect Conservation and Diversity, 5 (4). 298-311. 10.1111/j.1752-4598.2011.00161.x © 2011 The Authors. Insect Conservation and Diversity © 2011 The Royal Entomological Society This version available http://nora.nerc.ac.uk/18814/ NERC has developed NORA to enable users to access research outputs wholly or partially funded by NERC. Copyright and other rights for material on this site are retained by the rights owners. Users should read the terms and conditions of use of this material at http://nora.nerc.ac.uk/policies.html#access This document is the author’s final manuscript version of the journal article, incorporating any revisions agreed during the peer review process. Some differences between this and the publisher’s version remain. You are advised to consult the publisher’s version if you wish to cite from this article. The definitive version is available at http://onlinelibrary.wiley.com Contact CEH NORA team at [email protected] The NERC and CEH trademarks and logos (‘the Trademarks’) are registered trademarks of NERC in the UK and other countries, and may not be used without the prior written consent of the Trademark owner. Eight alien gallwasps in Britain 1 1 Range expansion and enemy recruitment by eight alien 2 gallwasp species in Britain 3 4 K. Schönrogge1, T. Begg1,2, R. Williams3, G. Melika4, Z. Randle5 & G.N. Stone2 5 6 1.