Analysis of the Rdr1 Gene Family in Different Rosaceae Genomes Reveals an Origin of an R- Gene Cluster After the Split of Rubeae Within the Rosoideae Subfamily
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MSRP Appendix E
Appendix E. Exotic Plant Species Reported from the South Florida Ecosystem. Community types are indicated where known Species High Pine Scrub Scrubby high pine Beach dune/ Coastal strand Maritime hammock Mesic temperate hammock Tropical hardwood Pine rocklands Scrubby flatwoods Mesic pine flatwoods Hydric pine flatwoods Dry prairie Cutthroat grass Wet prairie Freshwater marsh Seepage swamp Flowing water swamp Pond swamp Mangrove Salt marsh Abelmoschus esculentus Abrus precatorius X X X X X X X X X X X X Abutilon hirtum Abutilon theophrasti Acacia auriculiformis X X X X X X X X X Acacia retinoides Acacia sphaerocephala Acalypha alopecuroidea Acalypha amentacea ssp. wilkesiana Acanthospermum australe Acanthospermum hispidum Achyranthes aspera var. X aspera Achyranthes aspera var. pubescens Acmella pilosa Page E-1 Species High Pine Scrub Scrubby high pine Beach dune/ Coastal strand Maritime hammock Mesic temperate hammock Tropical hardwood Pine rocklands Scrubby flatwoods Mesic pine flatwoods Hydric pine flatwoods Dry prairie Cutthroat grass Wet prairie Freshwater marsh Seepage swamp Flowing water swamp Pond swamp Mangrove Salt marsh Acrocomia aculeata X Adenanthera pavonina X X Adiantum anceps X Adiantum caudatum Adiantum trapeziforme X Agave americana Agave angustifolia cv. X marginata Agave desmettiana Agave sisalana X X X X X X Agdestis clematidea X Ageratum conyzoides Ageratum houstonianum Aglaonema commutatum var. maculatum Ailanthus altissima Albizia julibrissin Albizia lebbeck X X X X X X X Albizia lebbeckoides Albizia procera Page -

South Carolina Wildflowers by Color and Season
SOUTH CAROLINA WILDFLOWERS *Chokeberry (Aronia arbutifolia) Silky Camellia (Stewartia malacodendron) BY COLOR AND SEASON Mountain Camelia (Stewartia ovata) Dwarf Witch Alder (Fothergilla gardenii) Revised 10/2007 by Mike Creel *Wild Plums (Prunus angustifolia, americana) 155 Cannon Trail Road Flatwoods Plum (Prunus umbellata) Lexington, SC 29073 *Shadberry or Sarvis Tree (Amelanchier arborea, obovata) Phone: (803) 359-2717 E-mail: [email protected] Fringe Tree (Chionanthus virginicus) Yellowwood Tree (Cladratis kentuckeana) Silverbell Tree (Halesia carolina, etc.) IDENTIFY PLANTS BY COLOR, THEN Evergreen Cherry Laurel (Prunus caroliniana) SEASON . Common ones in bold print. Hawthorn (Crataegus viridis, marshalli, etc.) Storax (Styrax americana, grandifolia) Wild Crabapple (Malus angustifolia) WHITE Wild Cherry (Prunus serotina) SPRING WHITE Dec. 1 to May 15 SUMMER WHITE May 15 to Aug. 7 *Atamasco Lily (Zephyranthes atamasco) *Swamp Spiderlily (Hymenocallis crassifolia) Carolina Anemone (Anemone caroliniana) Rocky Shoals Spiderlily (Hymenocallis coronaria) Lance-leaved Anemone (Anemone lancifolia) Colic Root (Aletris farinosa) Meadow Anemone (Anemone canadensis) Fly-Poison (Amianthium muscaetoxicum) American Wood Anemone (Anemone quinquefolia) Angelica (Angelica venosa) Wild Indigo (Baptisia bracteata) Ground Nut Vine (Apios americana) Sandwort (Arenaria caroliniana) Indian Hemp (Apocynum cannabium) American Bugbane (Cimicifuga americana) Sand Milkweed (Asclepias humistrata) Cohosh Bugbane (Cimicifuga racemosa) White Milkweed (Asclepias -
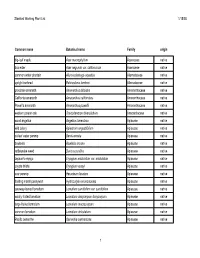
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Molecular Evidence for Hybrid Origin and Phenotypic Variation of Rosa Section Chinenses
G C A T T A C G G C A T genes Article Molecular Evidence for Hybrid Origin and Phenotypic Variation of Rosa Section Chinenses Chenyang Yang, Yujie Ma, Bixuan Cheng, Lijun Zhou, Chao Yu * , Le Luo, Huitang Pan and Qixiang Zhang Beijing Key Laboratory of Ornamental Plants Germplasm Innovation & Molecular Breeding, National Engineering Research Center for Floriculture, Beijing Laboratory of Urban and Rural Ecological Environment, Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Ministry of Education, School of Landscape Architecture, Beijing Forestry University, Beijing 100083, China; [email protected] (C.Y.); [email protected] (Y.M.); [email protected] (B.C.); [email protected] (L.Z.); [email protected] (L.L.); [email protected] (H.P.); [email protected] (Q.Z.) * Correspondence: [email protected] Received: 26 June 2020; Accepted: 18 August 2020; Published: 25 August 2020 Abstract: Rosa sect. Chinenses (Rosaceae) is an important parent of modern rose that is widely distributed throughout China and plays an important role in breeding and molecular biological research. R. sect. Chinenses has variable morphological traits and mixed germplasm. However, the taxonomic status and genetic background of sect. Chinenses varieties remain unclear. In this study, we collected germplasm resources from sect. Chinenses varieties with different morphological traits. Simple sequence repeat (SSR) markers, chloroplast markers, and single copy nuclear markers were used to explore the genetic background of these germplasm resources. We described the origin of hybridization of rose germplasm resources by combining different molecular markers. The results showed that the flower and hip traits of different species in R. -

Colonial Garden Plants
COLONIAL GARD~J~ PLANTS I Flowers Before 1700 The following plants are listed according to the names most commonly used during the colonial period. The botanical name follows for accurate identification. The common name was listed first because many of the people using these lists will have access to or be familiar with that name rather than the botanical name. The botanical names are according to Bailey’s Hortus Second and The Standard Cyclopedia of Horticulture (3, 4). They are not the botanical names used during the colonial period for many of them have changed drastically. We have been very cautious concerning the interpretation of names to see that accuracy is maintained. By using several references spanning almost two hundred years (1, 3, 32, 35) we were able to interpret accurately the names of certain plants. For example, in the earliest works (32, 35), Lark’s Heel is used for Larkspur, also Delphinium. Then in later works the name Larkspur appears with the former in parenthesis. Similarly, the name "Emanies" appears frequently in the earliest books. Finally, one of them (35) lists the name Anemones as a synonym. Some of the names are amusing: "Issop" for Hyssop, "Pum- pions" for Pumpkins, "Mushmillions" for Muskmellons, "Isquou- terquashes" for Squashes, "Cowslips" for Primroses, "Daffadown dillies" for Daffodils. Other names are confusing. Bachelors Button was the name used for Gomphrena globosa, not for Centaurea cyanis as we use it today. Similarly, in the earliest literature, "Marygold" was used for Calendula. Later we begin to see "Pot Marygold" and "Calen- dula" for Calendula, and "Marygold" is reserved for Marigolds. -

Rose Sampletext
A rose is a woody perennial flowering plant of the genus Rosa, in the • Synstylae – white, pink, and crimson flowered roses from all areas. family Rosaceae, or the flower it bears. There are over a hundred species and thousands of cultivars. They form a group of plants that can be erect shrubs, climbing or trailing with stems that are often Uses armed with sharp prickles. Flowers vary in size and shape and are Roses are best known as ornamental plants grown for their flowers in usually large and showy, in colours ranging from white through the garden and sometimes indoors. They have been also used for yellows and reds. Most species are native to Asia, with smaller numbers commercial perfumery and commercial cut flower crops. Some are used native to Europe, North America, and northwestern Africa. Species, as landscape plants, for hedging and for other utilitarian purposes such cultivars and hybrids are all widely grown for their beauty and often as game cover and slope stabilization. They also have minor medicinal are fragrant. Roses have acquired cultural significance in many uses. societies. Rose plants range in size from compact, miniature roses, to Ornamental plants climbers that can reach seven meters in height. Different species The majority of ornamental roses are hybrids that were bred for their hybridize easily, and this has been used in the development of the wide flowers. A few, mostly species roses are grown for attractive or scented range of garden roses. foliage (such as Rosa glauca and Rosa rubiginosa), ornamental thorns The name rose comes from French, itself from Latin rosa, which was (such as Rosa sericea) or for their showy fruit (such as Rosa moyesii). -

Resemblances and Disparities of Two Biotas Ziwei Zhang
Resemblances and Disparities of two Biotas A Comparison Study of Vascular Plant Biodiversity of two Locations in Uppsala and Beijing Ziwei Zhang Degree project in biology, Bachelor of science, 2013 Examensarbete i biologi 15 hp till kandidatexamen, 2013 Institutionen för biologisk grundutbildning och Avd för växtekologi och evolution, Uppsala universitet Handledare: Håkan Rydin Abstract This paper focuses on the flora distribution and difference in biodiversities of two chosen locations in Uppsala and Beijing, through inventorial and analytic methods. The factors that may cause the difference were also discussed from theoretical perspectives. Inventories of vascular plant species were carried out in two locations of the two cities. The collected species data were then grouped into families as well as life forms; and were compared with each other as well as with the statistics from the entire species pool in the chosen city. Both resemblances and disparities were found. The statistical analyses with Minitab supported the hypotheses that the floral compositions of these two locations differ to a great extent. Various factors such as climate, grazing, human impacts, historical reasons, precipitation, humidity and evolution, can account for the disparities. 2 Contents ABSTRACT ............................................................................................................................................................ 2 1. INTRODUCTION ............................................................................................................................................. -

Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B
Aliso: A Journal of Systematic and Evolutionary Botany Volume 29 | Issue 1 Article 4 2011 Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B. Harper Terra Peninsular, Coronado, California Sula Vanderplank Rancho Santa Ana Botanic Garden, Claremont, California Mark Dodero Recon Environmental Inc., San Diego, California Sergio Mata Terra Peninsular, Coronado, California Jorge Ochoa Long Beach City College, Long Beach, California Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Biodiversity Commons, Botany Commons, and the Ecology and Evolutionary Biology Commons Recommended Citation Harper, Alan B.; Vanderplank, Sula; Dodero, Mark; Mata, Sergio; and Ochoa, Jorge (2011) "Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 29: Iss. 1, Article 4. Available at: http://scholarship.claremont.edu/aliso/vol29/iss1/4 Aliso, 29(1), pp. 25–42 ’ 2011, Rancho Santa Ana Botanic Garden PLANTS OF THE COLONET REGION, BAJA CALIFORNIA, MEXICO, AND A VEGETATION MAPOF COLONET MESA ALAN B. HARPER,1 SULA VANDERPLANK,2 MARK DODERO,3 SERGIO MATA,1 AND JORGE OCHOA4 1Terra Peninsular, A.C., PMB 189003, Suite 88, Coronado, California 92178, USA ([email protected]); 2Rancho Santa Ana Botanic Garden, 1500 North College Avenue, Claremont, California 91711, USA; 3Recon Environmental Inc., 1927 Fifth Avenue, San Diego, California 92101, USA; 4Long Beach City College, 1305 East Pacific Coast Highway, Long Beach, California 90806, USA ABSTRACT The Colonet region is located at the southern end of the California Floristic Province, in an area known to have the highest plant diversity in Baja California. -

Letter Report (December 7, 2020)
Appendix B Biological Letter Report (December 7, 2020) STREET 605 THIRD 92024 CALIFORNIA ENCINITAS. F 760.632.0164 T 760.942.5147 December 7, 2020 11575 John R. Tschudin, Jr. Director – Design & Construction Encompass Health 9001 Liberty Parkway Birmingham, Alabama 35242 Subject: Biology Letter Report for Encompass Health Chula Vista, City of Chula Vista, California Dear Mr. Tschudin: This letter report provides an analysis of potential biological resource impacts associated with Encompass Health Chula Vista (proposed project) located in the City of Chula Vista (City), California (Assessor’s Parcel Number 644- 040-01-00). This biology letter report also includes a discussion of any potential biological resources that may be subject to regulation under the City of Chula Vista Multiple Species Conservation Program (MSCP) Subarea Plan (Subarea Plan) (City of Chula Vista 2003). Project Location The property (i.e., on-site; Assessor’s Parcel Number 644-040-01-00) occupies 9.79 acres and is located approximately 0.2 miles east of Interstate 805 between Main Street and Olympic Parkway (Figure 1, Project Location). The project also includes an off-site impact area of 0.22 acre located along the southeastern corner of the site where future utility connections may occur, making the total study area acreage for the project 10.01 acres. The site is located on Shinohara Lane accessed from Brandywine Avenue and is located on the U.S. Geological Service 7.5-minute series topographic Imperial Beach quadrangle map. The site exists within an urban portion of the City and is bound on the south and east by industrial buildings, to the west by single-family residences, and to the north by multi-family condominiums (Figure 2, Aerial Image). -

Literature Cited
Literature Cited Robert W. Kiger, Editor This is a consolidated list of all works cited in volume 9, whether as selected references, in text, or in nomenclatural contexts. In citations of articles, both here and in the taxonomic treatments, and also in nomenclatural citations, the titles of serials are rendered in the forms recommended in G. D. R. Bridson and E. R. Smith (1991), Bridson (2004), and Bridson and D. W. Brown (http://fmhibd.library.cmu.edu/fmi/iwp/cgi?-db=BPH_Online&-loadframes). When those forms are abbreviated, as most are, cross references to the corresponding full serial titles are interpolated here alphabetically by abbreviated form. In nomenclatural citations (only), book titles are rendered in the abbreviated forms recommended in F. A. Stafleu and R. S. Cowan (1976–1988) and Stafleu et al. (1992–2009). Here, those abbreviated forms are indicated parenthetically following the full citations of the corresponding works, and cross references to the full citations are interpolated in the list alphabetically by abbreviated form. Two or more works published in the same year by the same author or group of coauthors will be distinguished uniquely and consistently throughout all volumes of Flora of North America by lower-case letters (b, c, d, ...) suffixed to the date for the second and subsequent works in the set. The suffixes are assigned in order of editorial encounter and do not reflect chronological sequence of publication. The first work by any particular author or group from any given year carries the implicit date suffix “a”; thus, the sequence of explicit suffixes begins with “b”. -

Fresh Floral Resource Guide Seasonal Floral
Fresh Floral RESOURCE GUIDE SEASONAL FLORAL Everyday Spring Summer Fall Winter Alstroemeria Agapanthus Agapanthus Autumn Leaves Amaryllis Anthurium Amaryllis Amaranthus Chrysanthemum Anemone Aster Anemone Astilbe Dahlia Evergreen Bells of Ireland Cherry Blossom Cosmos Heather Heather Bupleurum Daffodil Dahlia Nerine Lily Muscari Calla Lily Dogwood Daisy Ranunculus Nerine Lily Carnation Forsythia Delphinium Seasonal Berries Poinsettia Craspedia Heather Garden Rose Sunflower Ranunculus Eryngium Hyacinth Gladiolus Tuberose Tulip Fiddlehead Fern Lilac Lady Mantle Waxflower Freesia Lily of the Valley Larkspur Gardenia Muscari Nerine Lily Gerbera Nerine Lily Scabiosa Gloriosa Lily Peony Snapdragon Hydrangea Ranunculus Tuberose Hypericum Sweet Pea Violet Iris Tuberose Zinnia Kermit’ Pompom Tulip Liatris Viburnum Lily Waxflower Limonium Lisianthus Lotus Pod Orchird Ornithogalum Arabicum Queen Anne’s Lace Rose Solidago Solidaster Spider Gerbera Statice Stephanotis Stock Trachelium Viking Mini Pompom BloomNet Fresh Floral Resource Guide 1 CARE & HANDLING Alstroemeria Alstroemeria is extremely ethylene sensitive. Remove any foliage that will go below the water line, as it deteriorates quickly. Then cut stems and place in a vase with room temperature water and floral food. Category: Basic Color: Yellow, Orange, White, Pink, Red, Lavender, Purple, Magenta, Peach, Bi Color Amaryllis Do not store amaryllis at below 41 degrees as this may discolor blooms. Amaryllis flowers damage very easily in bud or bloom stage. Allow space around the blooms in a bucket or design to prohibit damage. Category: Novelty Color: White, Pink, Red, Peach, Orange, Bi Color Anthurium An extremely long lasting Tropical flower with a vase life from 15 to 30 days (mini and small grades tend to have a shorter vase life). -

Plant Taxonomy: What's in a Name?
Episode: Thewwwwwwwwwwwwwwwwwwwwww Naturalists EXPLORING NORTH CAROLINA Plant Taxonomy: What’s in a Name? STANDARD COURSE OF STUDY CORRELATIONS: Biology, Goal 4: The learner will develop an understanding of the unity and diversity of life. 4.01 Analyze the classification of organisms. Biology, Goal 5: The learner will develop an understanding of the ecological relationships among organisms. 5.03 Assess human population and its impact on local ecosystems and global environments. INTRODUCTION TO LESSON: Students will investigate the usefulness of taxonomy by deciphering the scientific names of North Carolina plants identi fied by explorer and naturalist André Michaux. They will complete a graphic organ MATERIALS izer using reference manuals, field guides and/or Internet resources to help them w Copies of assignment understand the significance of scientific names. sheet, one per pair of students BACKGROUND FOR TEACHER: Many early European explorers came to the New World w Viewing Guides, one looking for fairly common plants that were considered valuable natural resources. per student André Michaux was among the brave botanists who endured harrowing conditions w Reference books (see traveling through the unexplored backwoods. The French government sent Michaux Additional Resources) to North Carolina in 1785 to look for timber to supplement France’s depleted stocks. Michaux exceeded this mission, eventually sending 60,000 plant specimens back PREPARATION to France. In the Carolinas alone, he named and identified close to 300 plants that w Make copies of Viewing were new to science. Guide and Assignment Sheets. w If students will not have classroom Internet access, engage f Ask students to name some reasons that plants are useful.