Efficient Methods to Calculate Partial Sphere Surface Areas for A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 11. Three Dimensional Analytic Geometry and Vectors
Chapter 11. Three dimensional analytic geometry and vectors. Section 11.5 Quadric surfaces. Curves in R2 : x2 y2 ellipse + =1 a2 b2 x2 y2 hyperbola − =1 a2 b2 parabola y = ax2 or x = by2 A quadric surface is the graph of a second degree equation in three variables. The most general such equation is Ax2 + By2 + Cz2 + Dxy + Exz + F yz + Gx + Hy + Iz + J =0, where A, B, C, ..., J are constants. By translation and rotation the equation can be brought into one of two standard forms Ax2 + By2 + Cz2 + J =0 or Ax2 + By2 + Iz =0 In order to sketch the graph of a quadric surface, it is useful to determine the curves of intersection of the surface with planes parallel to the coordinate planes. These curves are called traces of the surface. Ellipsoids The quadric surface with equation x2 y2 z2 + + =1 a2 b2 c2 is called an ellipsoid because all of its traces are ellipses. 2 1 x y 3 2 1 z ±1 ±2 ±3 ±1 ±2 The six intercepts of the ellipsoid are (±a, 0, 0), (0, ±b, 0), and (0, 0, ±c) and the ellipsoid lies in the box |x| ≤ a, |y| ≤ b, |z| ≤ c Since the ellipsoid involves only even powers of x, y, and z, the ellipsoid is symmetric with respect to each coordinate plane. Example 1. Find the traces of the surface 4x2 +9y2 + 36z2 = 36 1 in the planes x = k, y = k, and z = k. Identify the surface and sketch it. Hyperboloids Hyperboloid of one sheet. The quadric surface with equations x2 y2 z2 1. -

An Introduction to Topology the Classification Theorem for Surfaces by E
An Introduction to Topology An Introduction to Topology The Classification theorem for Surfaces By E. C. Zeeman Introduction. The classification theorem is a beautiful example of geometric topology. Although it was discovered in the last century*, yet it manages to convey the spirit of present day research. The proof that we give here is elementary, and its is hoped more intuitive than that found in most textbooks, but in none the less rigorous. It is designed for readers who have never done any topology before. It is the sort of mathematics that could be taught in schools both to foster geometric intuition, and to counteract the present day alarming tendency to drop geometry. It is profound, and yet preserves a sense of fun. In Appendix 1 we explain how a deeper result can be proved if one has available the more sophisticated tools of analytic topology and algebraic topology. Examples. Before starting the theorem let us look at a few examples of surfaces. In any branch of mathematics it is always a good thing to start with examples, because they are the source of our intuition. All the following pictures are of surfaces in 3-dimensions. In example 1 by the word “sphere” we mean just the surface of the sphere, and not the inside. In fact in all the examples we mean just the surface and not the solid inside. 1. Sphere. 2. Torus (or inner tube). 3. Knotted torus. 4. Sphere with knotted torus bored through it. * Zeeman wrote this article in the mid-twentieth century. 1 An Introduction to Topology 5. -
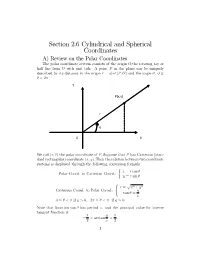
Section 2.6 Cylindrical and Spherical Coordinates
Section 2.6 Cylindrical and Spherical Coordinates A) Review on the Polar Coordinates The polar coordinate system consists of the origin O,the rotating ray or half line from O with unit tick. A point P in the plane can be uniquely described by its distance to the origin r = dist (P, O) and the angle µ, 0 µ < 2¼ : · Y P(x,y) r θ O X We call (r, µ) the polar coordinate of P. Suppose that P has Cartesian (stan- dard rectangular) coordinate (x, y) .Then the relation between two coordinate systems is displayed through the following conversion formula: x = r cos µ Polar Coord. to Cartesian Coord.: y = r sin µ ½ r = x2 + y2 Cartesian Coord. to Polar Coord.: y tan µ = ( p x 0 µ < ¼ if y > 0, 2¼ µ < ¼ if y 0. · · · Note that function tan µ has period ¼, and the principal value for inverse tangent function is ¼ y ¼ < arctan < . ¡ 2 x 2 1 So the angle should be determined by y arctan , if x > 0 xy 8 arctan + ¼, if x < 0 µ = > ¼ x > > , if x = 0, y > 0 < 2 ¼ , if x = 0, y < 0 > ¡ 2 > > Example 6.1. Fin:>d (a) Cartesian Coord. of P whose Polar Coord. is ¼ 2, , and (b) Polar Coord. of Q whose Cartesian Coord. is ( 1, 1) . 3 ¡ ¡ ³ So´l. (a) ¼ x = 2 cos = 1, 3 ¼ y = 2 sin = p3. 3 (b) r = p1 + 1 = p2 1 ¼ ¼ 5¼ tan µ = ¡ = 1 = µ = or µ = + ¼ = . 1 ) 4 4 4 ¡ 5¼ Since ( 1, 1) is in the third quadrant, we choose µ = so ¡ ¡ 4 5¼ p2, is Polar Coord. -

Surface Physics I and II
Surface physics I and II Lectures: Mon 12-14 ,Wed 12-14 D116 Excercises: TBA Lecturer: Antti Kuronen, [email protected] Exercise assistant: Ane Lasa, [email protected] Course homepage: http://www.physics.helsinki.fi/courses/s/pintafysiikka/ Objectives ● To study properties of surfaces of solid materials. ● The relationship between the composition and morphology of the surface and its mechanical, chemical and electronic properties will be dealt with. ● Technologically important field of surface and thin film growth will also be covered. Surface physics I 2012: 1. Introduction 1 Surface physics I and II ● Course in two parts ● Surface physics I (SPI) (530202) ● Period III, 5 ECTS points ● Basics of surface physics ● Surface physics II (SPII) (530169) ● Period IV, 5 ECTS points ● 'Special' topics in surface science ● Surface and thin film growth ● Nanosystems ● Computational methods in surface science ● You can take only SPI or both SPI and SPII Surface physics I 2012: 1. Introduction 2 How to pass ● Both courses: ● Final exam 50% ● Exercises 50% ● Exercises ● Return by ● email to [email protected] or ● on paper to course box on the 2nd floor of Physicum ● Return by (TBA) Surface physics I 2012: 1. Introduction 3 Table of contents ● Surface physics I ● Introduction: What is a surface? Why is it important? Basic concepts. ● Surface structure: Thermodynamics of surfaces. Atomic and electronic structure. ● Experimental methods for surface characterization: Composition, morphology, electronic properties. ● Surface physics II ● Theoretical and computational methods in surface science: Analytical models, Monte Carlo and molecular dynamics sumilations. ● Surface growth: Adsorption, desorption, surface diffusion. ● Thin film growth: Homoepitaxy, heteroepitaxy, nanostructures. -

Area, Volume and Surface Area
The Improving Mathematics Education in Schools (TIMES) Project MEASUREMENT AND GEOMETRY Module 11 AREA, VOLUME AND SURFACE AREA A guide for teachers - Years 8–10 June 2011 YEARS 810 Area, Volume and Surface Area (Measurement and Geometry: Module 11) For teachers of Primary and Secondary Mathematics 510 Cover design, Layout design and Typesetting by Claire Ho The Improving Mathematics Education in Schools (TIMES) Project 2009‑2011 was funded by the Australian Government Department of Education, Employment and Workplace Relations. The views expressed here are those of the author and do not necessarily represent the views of the Australian Government Department of Education, Employment and Workplace Relations. © The University of Melbourne on behalf of the international Centre of Excellence for Education in Mathematics (ICE‑EM), the education division of the Australian Mathematical Sciences Institute (AMSI), 2010 (except where otherwise indicated). This work is licensed under the Creative Commons Attribution‑NonCommercial‑NoDerivs 3.0 Unported License. http://creativecommons.org/licenses/by‑nc‑nd/3.0/ The Improving Mathematics Education in Schools (TIMES) Project MEASUREMENT AND GEOMETRY Module 11 AREA, VOLUME AND SURFACE AREA A guide for teachers - Years 8–10 June 2011 Peter Brown Michael Evans David Hunt Janine McIntosh Bill Pender Jacqui Ramagge YEARS 810 {4} A guide for teachers AREA, VOLUME AND SURFACE AREA ASSUMED KNOWLEDGE • Knowledge of the areas of rectangles, triangles, circles and composite figures. • The definitions of a parallelogram and a rhombus. • Familiarity with the basic properties of parallel lines. • Familiarity with the volume of a rectangular prism. • Basic knowledge of congruence and similarity. • Since some formulas will be involved, the students will need some experience with substitution and also with the distributive law. -

Surface Water
Chapter 5 SURFACE WATER Surface water originates mostly from rainfall and is a mixture of surface run-off and ground water. It includes larges rivers, ponds and lakes, and the small upland streams which may originate from springs and collect the run-off from the watersheds. The quantity of run-off depends upon a large number of factors, the most important of which are the amount and intensity of rainfall, the climate and vegetation and, also, the geological, geographi- cal, and topographical features of the area under consideration. It varies widely, from about 20 % in arid and sandy areas where the rainfall is scarce to more than 50% in rocky regions in which the annual rainfall is heavy. Of the remaining portion of the rainfall. some of the water percolates into the ground (see "Ground water", page 57), and the rest is lost by evaporation, transpiration and absorption. The quality of surface water is governed by its content of living organisms and by the amounts of mineral and organic matter which it may have picked up in the course of its formation. As rain falls through the atmo- sphere, it collects dust and absorbs oxygen and carbon dioxide from the air. While flowing over the ground, surface water collects silt and particles of organic matter, some of which will ultimately go into solution. It also picks up more carbon dioxide from the vegetation and micro-organisms and bacteria from the topsoil and from decaying matter. On inhabited watersheds, pollution may include faecal material and pathogenic organisms, as well as other human and industrial wastes which have not been properly disposed of. -

Riemann Surfaces
RIEMANN SURFACES AARON LANDESMAN CONTENTS 1. Introduction 2 2. Maps of Riemann Surfaces 4 2.1. Defining the maps 4 2.2. The multiplicity of a map 4 2.3. Ramification Loci of maps 6 2.4. Applications 6 3. Properness 9 3.1. Definition of properness 9 3.2. Basic properties of proper morphisms 9 3.3. Constancy of degree of a map 10 4. Examples of Proper Maps of Riemann Surfaces 13 5. Riemann-Hurwitz 15 5.1. Statement of Riemann-Hurwitz 15 5.2. Applications 15 6. Automorphisms of Riemann Surfaces of genus ≥ 2 18 6.1. Statement of the bound 18 6.2. Proving the bound 18 6.3. We rule out g(Y) > 1 20 6.4. We rule out g(Y) = 1 20 6.5. We rule out g(Y) = 0, n ≥ 5 20 6.6. We rule out g(Y) = 0, n = 4 20 6.7. We rule out g(C0) = 0, n = 3 20 6.8. 21 7. Automorphisms in low genus 0 and 1 22 7.1. Genus 0 22 7.2. Genus 1 22 7.3. Example in Genus 3 23 Appendix A. Proof of Riemann Hurwitz 25 Appendix B. Quotients of Riemann surfaces by automorphisms 29 References 31 1 2 AARON LANDESMAN 1. INTRODUCTION In this course, we’ll discuss the theory of Riemann surfaces. Rie- mann surfaces are a beautiful breeding ground for ideas from many areas of math. In this way they connect seemingly disjoint fields, and also allow one to use tools from different areas of math to study them. -

3D Modeling: Surfaces
CS 430/536 Computer Graphics I Overview • 3D model representations 3D Modeling: • Mesh formats Surfaces • Bicubic surfaces • Bezier surfaces Week 8, Lecture 16 • Normals to surfaces David Breen, William Regli and Maxim Peysakhov • Direct surface rendering Geometric and Intelligent Computing Laboratory Department of Computer Science Drexel University 1 2 http://gicl.cs.drexel.edu 1994 Foley/VanDam/Finer/Huges/Phillips ICG 3D Modeling Representing 3D Objects • 3D Representations • Exact • Approximate – Wireframe models – Surface Models – Wireframe – Facet / Mesh – Solid Models – Parametric • Just surfaces – Meshes and Polygon soups – Voxel/Volume models Surface – Voxel – Decomposition-based – Solid Model • Volume info • Octrees, voxels • CSG • Modeling in 3D – Constructive Solid Geometry (CSG), • BRep Breps and feature-based • Implicit Solid Modeling 3 4 Negatives when Representing 3D Objects Representing 3D Objects • Exact • Approximate • Exact • Approximate – Complex data structures – Lossy – Precise model of – A discretization of – Expensive algorithms – Data structure sizes can object topology the 3D object – Wide variety of formats, get HUGE, if you want each with subtle nuances good fidelity – Mathematically – Use simple – Hard to acquire data – Easy to break (i.e. cracks represent all primitives to – Translation required for can appear) rendering – Not good for certain geometry model topology applications • Lots of interpolation and and geometry guess work 5 6 1 Positives when Exact Representations Representing 3D Objects • Exact -

Visual Cortex: Looking Into a Klein Bottle Nicholas V
776 Dispatch Visual cortex: Looking into a Klein bottle Nicholas V. Swindale Arguments based on mathematical topology may help work which suggested that many properties of visual cortex in understanding the organization of topographic maps organization might be a consequence of mapping a five- in the cerebral cortex. dimensional stimulus space onto a two-dimensional surface as continuously as possible. Recent experimental results, Address: Department of Ophthalmology, University of British Columbia, 2550 Willow Street, Vancouver, V5Z 3N9, British which I shall discuss here, add to this complexity because Columbia, Canada. they show that a stimulus attribute not considered in the theoretical studies — direction of motion — is also syst- Current Biology 1996, Vol 6 No 7:776–779 ematically mapped on the surface of the cortex. This adds © Current Biology Ltd ISSN 0960-9822 to the evidence that continuity is an important, though not overriding, organizational principle in the cortex. I shall The English neurologist Hughlings Jackson inferred the also discuss a demonstration that certain receptive-field presence of a topographic map of the body musculature in properties, which may be indirectly related to direction the cerebral cortex more than a century ago, from his selectivity, can be represented as positions in a non- observations of the orderly progressions of seizure activity Euclidian space with a topology known to mathematicians across the body during epilepsy. Topographic maps of one as a Klein bottle. First, however, it is appropriate to cons- kind or another are now known to be a ubiquitous feature ider the experimental data. of cortical organization, at least in the primary sensory and motor areas. -

Analytic Geometry
STATISTIC ANALYTIC GEOMETRY SESSION 3 STATISTIC SESSION 3 Session 3 Analytic Geometry Geometry is all about shapes and their properties. If you like playing with objects, or like drawing, then geometry is for you! Geometry can be divided into: Plane Geometry is about flat shapes like lines, circles and triangles ... shapes that can be drawn on a piece of paper Solid Geometry is about three dimensional objects like cubes, prisms, cylinders and spheres Point, Line, Plane and Solid A Point has no dimensions, only position A Line is one-dimensional A Plane is two dimensional (2D) A Solid is three-dimensional (3D) Plane Geometry Plane Geometry is all about shapes on a flat surface (like on an endless piece of paper). 2D Shapes Activity: Sorting Shapes Triangles Right Angled Triangles Interactive Triangles Quadrilaterals (Rhombus, Parallelogram, etc) Rectangle, Rhombus, Square, Parallelogram, Trapezoid and Kite Interactive Quadrilaterals Shapes Freeplay Perimeter Area Area of Plane Shapes Area Calculation Tool Area of Polygon by Drawing Activity: Garden Area General Drawing Tool Polygons A Polygon is a 2-dimensional shape made of straight lines. Triangles and Rectangles are polygons. Here are some more: Pentagon Pentagra m Hexagon Properties of Regular Polygons Diagonals of Polygons Interactive Polygons The Circle Circle Pi Circle Sector and Segment Circle Area by Sectors Annulus Activity: Dropping a Coin onto a Grid Circle Theorems (Advanced Topic) Symbols There are many special symbols used in Geometry. Here is a short reference for you: -

Motion on a Torus Kirk T
Motion on a Torus Kirk T. McDonald Joseph Henry Laboratories, Princeton University, Princeton, NJ 08544 (October 21, 2000) 1 Problem Find the frequency of small oscillations about uniform circular motion of a point mass that is constrained to move on the surface of a torus (donut) of major radius a and minor radius b whose axis is vertical. 2 Solution 2.1 Attempt at a Quick Solution Circular orbits are possible in both horizontal and vertical planes, but in the presence of gravity, motion in vertical orbits will be at a nonuniform velocity. Hence, we restrict our attention to orbits in horizontal planes. We use a cylindrical coordinate system (r; θ; z), with the origin at the center of the torus and the z axis vertically upwards, as shown below. 1 A point on the surface of the torus can also be described by two angular coordinates, one of which is the azimuth θ in the cylindrical coordinate system. The other angle we define as Á measured with respect to the plane z = 0 in a vertical plane that contains the point as well as the axis, as also shown in the figure above. We seek motion at constant angular velocity Ω about the z axis, which suggests that we consider a frame that rotates with this angular velocity. In this frame, the particle (whose mass we take to be unity) is at rest at angle Á0, and is subject to the downward force of 2 gravity g and the outward centrifugal force Ω (a + b cos Á0), as shown in the figure below. -

Surface Topology
2 Surface topology 2.1 Classification of surfaces In this second introductory chapter, we change direction completely. We dis- cuss the topological classification of surfaces, and outline one approach to a proof. Our treatment here is almost entirely informal; we do not even define precisely what we mean by a ‘surface’. (Definitions will be found in the following chapter.) However, with the aid of some more sophisticated technical language, it not too hard to turn our informal account into a precise proof. The reasons for including this material here are, first, that it gives a counterweight to the previous chapter: the two together illustrate two themes—complex analysis and topology—which run through the study of Riemann surfaces. And, second, that we are able to introduce some more advanced ideas that will be taken up later in the book. The statement of the classification of closed surfaces is probably well known to many readers. We write down two families of surfaces g, h for integers g ≥ 0, h ≥ 1. 2 2 The surface 0 is the 2-sphere S . The surface 1 is the 2-torus T .For g ≥ 2, we define the surface g by taking the ‘connected sum’ of g copies of the torus. In general, if X and Y are (connected) surfaces, the connected sum XY is a surface constructed as follows (Figure 2.1). We choose small discs DX in X and DY in Y and cut them out to get a pair of ‘surfaces-with- boundaries’, coresponding to the circle boundaries of DX and DY.