Chemical Tools and Visualization Software for Computational Science
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

GPAW, Gpus, and LUMI
GPAW, GPUs, and LUMI Martti Louhivuori, CSC - IT Center for Science Jussi Enkovaara GPAW 2021: Users and Developers Meeting, 2021-06-01 Outline LUMI supercomputer Brief history of GPAW with GPUs GPUs and DFT Current status Roadmap LUMI - EuroHPC system of the North Pre-exascale system with AMD CPUs and GPUs ~ 550 Pflop/s performance Half of the resources dedicated to consortium members Programming for LUMI Finland, Belgium, Czechia, MPI between nodes / GPUs Denmark, Estonia, Iceland, HIP and OpenMP for GPUs Norway, Poland, Sweden, and how to use Python with AMD Switzerland GPUs? https://www.lumi-supercomputer.eu GPAW and GPUs: history (1/2) Early proof-of-concept implementation for NVIDIA GPUs in 2012 ground state DFT and real-time TD-DFT with finite-difference basis separate version for RPA with plane-waves Hakala et al. in "Electronic Structure Calculations on Graphics Processing Units", Wiley (2016), https://doi.org/10.1002/9781118670712 PyCUDA, cuBLAS, cuFFT, custom CUDA kernels Promising performance with factor of 4-8 speedup in best cases (CPU node vs. GPU node) GPAW and GPUs: history (2/2) Code base diverged from the main branch quite a bit proof-of-concept implementation had lots of quick and dirty hacks fixes and features were pulled from other branches and patches no proper unit tests for GPU functionality active development stopped soon after publications Before development re-started, code didn't even work anymore on modern GPUs without applying a few small patches Lesson learned: try to always get new functionality to the -

MODELLER 10.1 Manual
MODELLER A Program for Protein Structure Modeling Release 10.1, r12156 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/03/12 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

Free and Open Source Software for Computational Chemistry Education
Free and Open Source Software for Computational Chemistry Education Susi Lehtola∗,y and Antti J. Karttunenz yMolecular Sciences Software Institute, Blacksburg, Virginia 24061, United States zDepartment of Chemistry and Materials Science, Aalto University, Espoo, Finland E-mail: [email protected].fi Abstract Long in the making, computational chemistry for the masses [J. Chem. Educ. 1996, 73, 104] is finally here. We point out the existence of a variety of free and open source software (FOSS) packages for computational chemistry that offer a wide range of functionality all the way from approximate semiempirical calculations with tight- binding density functional theory to sophisticated ab initio wave function methods such as coupled-cluster theory, both for molecular and for solid-state systems. By their very definition, FOSS packages allow usage for whatever purpose by anyone, meaning they can also be used in industrial applications without limitation. Also, FOSS software has no limitations to redistribution in source or binary form, allowing their easy distribution and installation by third parties. Many FOSS scientific software packages are available as part of popular Linux distributions, and other package managers such as pip and conda. Combined with the remarkable increase in the power of personal devices—which rival that of the fastest supercomputers in the world of the 1990s—a decentralized model for teaching computational chemistry is now possible, enabling students to perform reasonable modeling on their own computing devices, in the bring your own device 1 (BYOD) scheme. In addition to the programs’ use for various applications, open access to the programs’ source code also enables comprehensive teaching strategies, as actual algorithms’ implementations can be used in teaching. -

User Guide of Computational Facilities
Physical Chemistry Unit Departament de Quimica User Guide of Computational Facilities Beta Version System Manager: Marc Noguera i Julian January 20, 2009 Contents 1 Introduction 1 2 Computer resources 2 3 Some Linux tools 2 4 The Cluster 2 4.1 Frontend servers . 2 4.2 Filesystems . 3 4.3 Computational nodes . 3 4.4 User’s environment . 4 4.5 Disk space quota . 4 4.6 Data Backup . 4 4.7 Generation of SSH keys for automatic SSH login . 4 4.8 Submitting jobs to the cluster . 5 4.8.1 Typical queue system commands . 6 4.8.2 HomeMade scripts . 6 4.8.3 Create your own submit script . 6 4.8.4 Submitting Parallel Calculations . 7 5 Available Software 9 5.1 Chemisty software . 9 5.2 Development Software . 10 5.3 How to use the software . 11 5.4 Non-default environments . 11 6 Your Linux Workstation 11 6.1 Disk space on your workstation . 12 6.2 Workstation Data Backup . 12 6.3 Running virtual Windows XP . 13 6.3.1 The Windows XP Environment . 13 6.4 Submitting calculations to your dekstop . 13 6.5 Network dependent structure . 14 7 Your WindowsXP Workstation 14 7.1 Workstation Data Backup . 14 8 Frequently asked questions 14 1 Introduction This User Guide is aimed to the standard user of the computer facilities in the ”Unitat de Quimica Fisica” in the Universitat Autnoma de Barcelona. It is not assumed that you have a knowledge of UNIX/Linux. However, you are 1 encouraged to take a close look to the linux guides that will be pointed out. -

MODELLER 10.0 Manual
MODELLER A Program for Protein Structure Modeling Release 10.0, r12005 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/01/25 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

Open Babel Documentation Release 2.3.1
Open Babel Documentation Release 2.3.1 Geoffrey R Hutchison Chris Morley Craig James Chris Swain Hans De Winter Tim Vandermeersch Noel M O’Boyle (Ed.) December 05, 2011 Contents 1 Introduction 3 1.1 Goals of the Open Babel project ..................................... 3 1.2 Frequently Asked Questions ....................................... 4 1.3 Thanks .................................................. 7 2 Install Open Babel 9 2.1 Install a binary package ......................................... 9 2.2 Compiling Open Babel .......................................... 9 3 obabel and babel - Convert, Filter and Manipulate Chemical Data 17 3.1 Synopsis ................................................. 17 3.2 Options .................................................. 17 3.3 Examples ................................................. 19 3.4 Differences between babel and obabel .................................. 21 3.5 Format Options .............................................. 22 3.6 Append property values to the title .................................... 22 3.7 Filtering molecules from a multimolecule file .............................. 22 3.8 Substructure and similarity searching .................................. 25 3.9 Sorting molecules ............................................ 25 3.10 Remove duplicate molecules ....................................... 25 3.11 Aliases for chemical groups ....................................... 26 4 The Open Babel GUI 29 4.1 Basic operation .............................................. 29 4.2 Options ................................................. -

Open Data, Open Source, and Open Standards in Chemistry: the Blue Obelisk Five Years On" Journal of Cheminformatics Vol
Oral Roberts University Digital Showcase College of Science and Engineering Faculty College of Science and Engineering Research and Scholarship 10-14-2011 Open Data, Open Source, and Open Standards in Chemistry: The lueB Obelisk five years on Andrew Lang Noel M. O'Boyle Rajarshi Guha National Institutes of Health Egon Willighagen Maastricht University Samuel Adams See next page for additional authors Follow this and additional works at: http://digitalshowcase.oru.edu/cose_pub Part of the Chemistry Commons Recommended Citation Andrew Lang, Noel M O'Boyle, Rajarshi Guha, Egon Willighagen, et al.. "Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on" Journal of Cheminformatics Vol. 3 Iss. 37 (2011) Available at: http://works.bepress.com/andrew-sid-lang/ 19/ This Article is brought to you for free and open access by the College of Science and Engineering at Digital Showcase. It has been accepted for inclusion in College of Science and Engineering Faculty Research and Scholarship by an authorized administrator of Digital Showcase. For more information, please contact [email protected]. Authors Andrew Lang, Noel M. O'Boyle, Rajarshi Guha, Egon Willighagen, Samuel Adams, Jonathan Alvarsson, Jean- Claude Bradley, Igor Filippov, Robert M. Hanson, Marcus D. Hanwell, Geoffrey R. Hutchison, Craig A. James, Nina Jeliazkova, Karol M. Langner, David C. Lonie, Daniel M. Lowe, Jerome Pansanel, Dmitry Pavlov, Ola Spjuth, Christoph Steinbeck, Adam L. Tenderholt, Kevin J. Theisen, and Peter Murray-Rust This article is available at Digital Showcase: http://digitalshowcase.oru.edu/cose_pub/34 Oral Roberts University From the SelectedWorks of Andrew Lang October 14, 2011 Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on Andrew Lang Noel M O'Boyle Rajarshi Guha, National Institutes of Health Egon Willighagen, Maastricht University Samuel Adams, et al. -

Molecular Docking Studies of a Cyclic Octapeptide-Cyclosaplin from Sandalwood
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 11 June 2019 doi:10.20944/preprints201906.0091.v1 Peer-reviewed version available at Biomolecules 2019, 9; doi:10.3390/biom9040123 Molecular Docking Studies of a Cyclic Octapeptide-Cyclosaplin from Sandalwood Abheepsa Mishra1, 2,* and Satyahari Dey1 1Plant Biotechnology Laboratory, Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur-721302, West Bengal, India; [email protected] (A.M.); [email protected] (S.D.) 2Department of Internal Medicine, The University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX 75390, USA *Correspondence: [email protected]; Tel.:+1(518) 881-9196 Abstract Natural products from plants such as, chemopreventive agents attract huge attention because of their low toxicity and high specificity. The rational drug design in combination with structure based modeling and rapid screening methods offer significant potential for identifying and developing lead anticancer molecules. Thus, the molecular docking method plays an important role in screening a large set of molecules based on their free binding energies and proposes structural hypotheses of how the molecules can inhibit the target. Several peptide based therapeutics have been developed to combat several health disorders including cancers, metabolic disorders, heart-related, and infectious diseases. Despite the discovery of hundreds of such therapeutic peptides however, only few peptide-based drugs have made it to the market. Moreover, until date the activities of cyclic peptides towards molecular targets such as protein kinases, proteases, and apoptosis related proteins have never been explored. In this study we explore the in silico kinase and protease inhibitor potentials of cyclosaplin as well as study the interactions of cyclosaplin with other cancer-related proteins. -
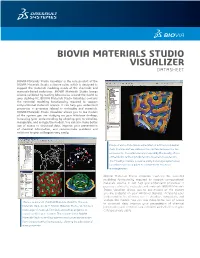
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

In Quantum Chemistry
http://www.cca-forum.org Computational Quality of Service (CQoS) in Quantum Chemistry Joseph Kenny1, Kevin Huck2, Li Li3, Lois Curfman McInnes3, Heather Netzloff4, Boyana Norris3, Meng-Shiou Wu4, Alexander Gaenko4 , and Hirotoshi Mori5 1Sandia National Laboratories, 2University of Oregon, 3Argonne National Laboratory, 4Ames Laboratory, 5Ochanomizu University, Japan This work is a collaboration among participants in the SciDAC Center for Technology for Advanced Scientific Component Software (TASCS), Performance Engineering Research Institute (PERI), Quantum Chemistry Science Application Partnership (QCSAP), and the Tuning and Analysis Utilities (TAU) group at the University of Oregon. Quantum Chemistry and the CQoS in Quantum Chemistry: Motivation and Approach Common Component Architecture (CCA) Motivation: CQoS Approach: CCA Overview: • QCSAP Challenges: How, during runtime, can we make the best choices • Overall: Develop infrastructure for dynamic component adaptivity, i.e., • The CCA Forum provides a specification and software tools for the for reliability, accuracy, and performance of interoperable quantum composing, substituting, and reconfiguring running CCA component development of high-performance components. chemistry components based on NWChem, MPQC, and GAMESS? applications in response to changing conditions – Performance, accuracy, mathematical consistency, reliability, etc. • Components = Composition – When several QC components provide the same functionality, what • Approach: Develop CQoS tools for – A component is a unit -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12