KLF2 and KLF4 Control Endothelial Identity and Vascular Integrity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microglia Emerge from Erythromyeloid Precursors Via Pu.1- and Irf8-Dependent Pathways
ART ic LE S Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways Katrin Kierdorf1,2, Daniel Erny1, Tobias Goldmann1, Victor Sander1, Christian Schulz3,4, Elisa Gomez Perdiguero3,4, Peter Wieghofer1,2, Annette Heinrich5, Pia Riemke6, Christoph Hölscher7,8, Dominik N Müller9, Bruno Luckow10, Thomas Brocker11, Katharina Debowski12, Günter Fritz1, Ghislain Opdenakker13, Andreas Diefenbach14, Knut Biber5,15, Mathias Heikenwalder16, Frederic Geissmann3,4, Frank Rosenbauer6 & Marco Prinz1,17 Microglia are crucial for immune responses in the brain. Although their origin from the yolk sac has been recognized for some time, their precise precursors and the transcription program that is used are not known. We found that mouse microglia were derived from primitive c-kit+ erythromyeloid precursors that were detected in the yolk sac as early as 8 d post conception. + lo − + − + These precursors developed into CD45 c-kit CX3CR1 immature (A1) cells and matured into CD45 c-kit CX3CR1 (A2) cells, as evidenced by the downregulation of CD31 and concomitant upregulation of F4/80 and macrophage colony stimulating factor receptor (MCSF-R). Proliferating A2 cells became microglia and invaded the developing brain using specific matrix metalloproteinases. Notably, microgliogenesis was not only dependent on the transcription factor Pu.1 (also known as Sfpi), but also required Irf8, which was vital for the development of the A2 population, whereas Myb, Id2, Batf3 and Klf4 were not required. Our data provide cellular and molecular insights into the origin and development of microglia. Microglia are the tissue macrophages of the brain and scavenge dying have the ability to give rise to microglia and macrophages in vitro cells, pathogens and molecules using pattern recognition receptors and in vivo under defined conditions. -

KLF2 Induced
UvA-DARE (Digital Academic Repository) The transcription factor KLF2 in vascular biology Boon, R.A. Publication date 2008 Link to publication Citation for published version (APA): Boon, R. A. (2008). The transcription factor KLF2 in vascular biology. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:23 Sep 2021 Supplementary data: Genes induced by KLF2 Dekker et al. LocusLink Accession Gene Sequence Description Fold p-value ID number symbol change (FDR) 6654 AK022099 SOS1 cDNA FLJ12037 fis, clone HEMBB1001921. 100.00 5.9E-09 56999 AF086069 ADAMTS9 full length insert cDNA clone YZ35C05. 100.00 1.2E-09 6672 AF085934 SP100 full length insert cDNA clone YR57D07. 100.00 6.7E-13 9031 AF132602 BAZ1B Williams Syndrome critical region WS25 mRNA, partial sequence. -

Angiogenic Patterning by STEEL, an Endothelial-Enriched Long
Angiogenic patterning by STEEL, an endothelial- enriched long noncoding RNA H. S. Jeffrey Mana,b, Aravin N. Sukumara,b, Gabrielle C. Lamc,d, Paul J. Turgeonb,e, Matthew S. Yanb,f, Kyung Ha Kub,e, Michelle K. Dubinskya,b, J. J. David Hob,f, Jenny Jing Wangb,e, Sunit Dasg,h, Nora Mitchelli, Peter Oettgeni, Michael V. Seftonc,d,j, and Philip A. Marsdena,b,e,f,1 aInstitute of Medical Science, University of Toronto, Toronto, ON M5S 1A8, Canada; bKeenan Research Centre for Biomedical Science in the Li Ka Shing Knowledge Institute, St. Michael’s Hospital, University of Toronto, Toronto, ON M5B 1T8, Canada; cDonnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON M5S 3E2, Canada; dInstitute of Biomaterials and Biomedical Engineering, University of Toronto, Toronto, ON M5S 3G9, Canada; eDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON M5S 1A8, Canada; fDepartment of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada; gArthur and Sonia Labatt Brain Tumour Research Institute, Hospital for SickKids, University of Toronto, Toronto, ON M5G 1X8, Canada; hDivision of Neurosurgery and Keenan Research Centre for Biomedical Science, St. Michael’s Hospital, University of Toronto, Toronto, ON M5B 1W8, Canada; iDepartment of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02115; and jDepartment of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada Edited by Napoleone Ferrara, University of California, San Diego, La Jolla, CA, and approved January 24, 2018 (received for review August 28, 2017) Endothelial cell (EC)-enriched protein coding genes, such as endothelial formation in vitro and blood vessel formation in vivo. -

A Dual Cis-Regulatory Code Links IRF8 to Constitutive and Inducible Gene Expression in Macrophages
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages Alessandra Mancino,1,3 Alberto Termanini,1,3 Iros Barozzi,1 Serena Ghisletti,1 Renato Ostuni,1 Elena Prosperini,1 Keiko Ozato,2 and Gioacchino Natoli1 1Department of Experimental Oncology, European Institute of Oncology (IEO), 20139 Milan, Italy; 2Laboratory of Molecular Growth Regulation, Genomics of Differentiation Program, National Institute of Child Health and Human Development (NICHD), National Institutes of Health, Bethesda, Maryland 20892, USA The transcription factor (TF) interferon regulatory factor 8 (IRF8) controls both developmental and inflammatory stimulus-inducible genes in macrophages, but the mechanisms underlying these two different functions are largely unknown. One possibility is that these different roles are linked to the ability of IRF8 to bind alternative DNA sequences. We found that IRF8 is recruited to distinct sets of DNA consensus sequences before and after lipopolysaccharide (LPS) stimulation. In resting cells, IRF8 was mainly bound to composite sites together with the master regulator of myeloid development PU.1. Basal IRF8–PU.1 binding maintained the expression of a broad panel of genes essential for macrophage functions (such as microbial recognition and response to purines) and contributed to basal expression of many LPS-inducible genes. After LPS stimulation, increased expression of IRF8, other IRFs, and AP-1 family TFs enabled IRF8 binding to thousands of additional regions containing low-affinity multimerized IRF sites and composite IRF–AP-1 sites, which were not premarked by PU.1 and did not contribute to the basal IRF8 cistrome. -

Inhibition of Adipocyte Differentiation by Rorα
FEBS Letters 583 (2009) 2031–2036 journal homepage: www.FEBSLetters.org Inhibition of adipocyte differentiation by RORa Hélène Duez a,b,c,1, Christian Duhem a,b,c,1, Saara Laitinen a,b,c, Prashant S. Patole a,b,c, Mouaadh Abdelkarim a,b,c, Brigitte Bois-Joyeux d, Jean-Louis Danan d, Bart Staels a,b,c,* a Institut Pasteur de Lille, Département d’Athérosclérose, Lille F-59019, France b INSERM UMR 545, Lille F-59019, France c Université Lille Nord de France, Faculté des Sciences Pharmaceutiques et Biologiques et Faculté de Médecine, Lille F-59006, France d Centre National de la Recherche Scientifique FRE3210, Faculté de Médecine René Descartes Paris 5, 75015 Paris, France article info abstract Article history: Here we show that gene expression of the nuclear receptor RORa is induced during adipogenesis, Received 8 April 2009 with RORa4 being the most abundantly expressed isoform in human and murine adipose tissue. Revised 27 April 2009 Over-expression of RORa4 in 3T3-L1 cells impairs adipogenesis as shown by the decreased expres- Accepted 8 May 2009 sion of adipogenic markers and lipid accumulation, accompanied by decreased free fatty acid and Available online 18 May 2009 glucose uptake. By contrast, mouse embryonic fibroblasts from staggerer mice, which carry a muta- Edited by Robert Barouki tion in the RORa gene, differentiate more efficiently into mature adipocytes compared to wild-type cells, a phenotype which is reversed by ectopic RORa4 restoration. Ó 2009 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved. Keywords: Adipogenesis RORa Nuclear receptors RAR-related orphan receptors (RORs) 1. -

Genequery™ Human Stem Cell Transcription Factors Qpcr Array
GeneQuery™ Human Stem Cell Transcription Factors qPCR Array Kit (GQH-SCT) Catalog #GK125 Product Description ScienCell's GeneQuery™ Human Stem Cell Transcription Factors qPCR Array Kit (GQH-SCT) surveys a panel of 88 transcription factors related to stem cell maintenance and differentiation. Transcription factors are proteins that can regulate target gene transcription level by binding to specific regions of the genome known as enhancers or silencers. Brief examples of how genes may be grouped according to their functions are shown below: • Pluripotency transcription factors: NANOG, POU5F1, SOX2 • Embryonic development: EOMES, FOXC2/D3/O1, HOXA9/A10, KLF2/5, LIN28B, MAX, PITX2, SMAD1, TBX5, ZIC1 • Germ layer formation and differentiation: FOXA2, GATA6, HAND1, ISL1, KLF4, SMAD2, SOX9 • Organ morphogenesis: EZH2, HOXA11/B3/B5/C9, MSX2, MYC, PAX5/8, VDR • Angiogenesis: CDX2, JUN, NR2F2, RUNX1, WT1 • Hematopoiesis and osteogenesis: EGR3, ESR1, GATA1/2/3, GLI2, NFATC1, PAX9, RB1, SOX6, SP1, STAT1 • Neural stem cell maintenance and differentiation: ATF2, CREB1, FOSB, FOXO3, HES1, HOXD10, MCM2/7, MEF2C, NEUROD1/G1, OLIG1/2, PAX3/6, POU4F1, PPARG, SMAD3/4, SNAI1, STAT3 Note : all gene names follow their official symbols by the Human Genome Organization Gene Nomenclature Committee (HGNC). GeneQuery™ qPCR array kits are qPCR ready in a 96-well plate format, with each well containing one primer set that can specifically recognize and efficiently amplify a target gene's cDNA. The carefully designed primers ensure that: (i) the optimal annealing temperature in qPCR analysis is 65°C (with 2 mM Mg 2+ , and no DMSO); (ii) the primer set recognizes all known transcript variants of target gene, unless otherwise indicated; and (iii) only one gene is amplified. -

Regulation of T Follicular Helper Cells by ICOS
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 26 Editorial Regulation of T follicular helper cells by ICOS Andreas Hutloff T follicular helper (TFH) cells are gatekeepers of the (OPN-i), which facilitates nuclear translocation [3]. In the humoral immune response. Without help from this CD4+ nucleus, OPN-i dimerizes with Bcl-6 and protects it from T cell subset, B cells cannot differentiate into high-affinity proteasomal degradation. However, this OPN-i and the memory B cells and antibody-producing long-lived plasma above Klf2 pathway do act at different times. Upon ICOS cells which are the basis of protective immune responses. signaling blockade, Klf2 is upregulated within a few hours At the same time, dysregulated TFH cell responses are and TFH cells lose their typical homing receptor pattern in causative for many autoimmune disorders (reviewed in less than 24 hours [6]. In contrast, complete degradation [1]). The inducible T cell costimulator ICOS, which is of Bcl-6 takes several days [3, 6]. Therefore, the ICOS - structurally and functionally related to CD28, has been Klf2 axis first leads to emigration of TFH cells out of the known for many years as an important regulator of TFH B cell follicle (and thereby to a loss of function), whereas cells. ICOS knock-out mice as well as ICOS-deficient the osteopontin pathway acts as a second strike pathway patients have only few TFH cells and very small germinal eliminating the lineage-defining transcription factor Bcl-6. centers upon immunization, which results in severely Another important finding especially for potential compromised antigen-specific immunoglobulin levels therapeutic applications is that ICOS and the structurally and the phenotype of common variable immunodeficiency related CD28 molecule act in different phases of TFH cell in humans (reviewed in [2]). -

Beta Cell Adaptation to Pregnancy Requires Prolactin Action on Both
www.nature.com/scientificreports OPEN Beta cell adaptation to pregnancy requires prolactin action on both beta and non‑beta cells Vipul Shrivastava1, Megan Lee1, Daniel Lee1, Marle Pretorius1, Bethany Radford1, Guneet Makkar1 & Carol Huang1,2,3* Pancreatic islets adapt to insulin resistance of pregnancy by up regulating β‑cell mass and increasing insulin secretion. Previously, using a transgenic mouse with global, heterozygous deletion of prolactin receptor (Prlr+/−), we found Prlr signaling is important for this adaptation. However, since Prlr is expressed in tissues outside of islets as well as within islets and prolactin signaling afects β‑cell development, to understand β‑cell‑specifc efect of prolactin signaling in pregnancy, we generated a transgenic mouse with an inducible conditional deletion of Prlr from β‑cells. Here, we found that β‑cell‑specifc Prlr reduction in adult mice led to elevated blood glucose, lowed β‑cell mass and blunted in vivo glucose‑stimulated insulin secretion during pregnancy. When we compared gene expression profle of islets from transgenic mice with global (Prlr+/−) versus β‑cell‑specifc Prlr reduction (βPrlR+/−), we found 95 diferentially expressed gene, most of them down regulated in the Prlr+/− mice in comparison to the βPrlR+/− mice, and many of these genes regulate apoptosis, synaptic vesicle function and neuronal development. Importantly, we found that islets from pregnant Prlr+/− mice are more vulnerable to glucolipotoxicity‑induced apoptosis than islets from pregnant βPrlR+/− mice. These observations suggest that down regulation of prolactin action during pregnancy in non‑β‑cells secondarily and negatively afect β‑cell gene expression, and increased β‑cell susceptibility to external insults. -

Krüppel-Like Factor 4 Promotes Survival and Expansion in Acute Myeloid Leukemia Cells
www.oncotarget.com Oncotarget, 2021, Vol. 12, (No. 4), pp: 255-267 Research Paper Krüppel-like factor 4 promotes survival and expansion in acute myeloid leukemia cells Andrew Henry Lewis1, Cory Seth Bridges1, Viraaj Singh Punia1, Abraham Fausto Jornada Cooper1,2, Monica Puppi1 and H. Daniel Lacorazza1 1Department of Pathology & Immunology, Baylor College of Medicine, Texas Children’s Hospital, Houston, TX 77030, USA 2SMART Program at Baylor College of Medicine Houston, Houston, TX 77030, USA Correspondence to: H. Daniel Lacorazza, email: [email protected] Keywords: acute myeloid leukemia; KLF4; cancer; gene editing; cell growth Received: November 20, 2020 Accepted: January 19, 2021 Published: February 16, 2021 Copyright: © 2021 Lewis et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Acute myeloid leukemia (AML) is an aggressive hematological malignancy of the bone marrow that affects mostly elderly adults. Alternative therapies are needed for AML patients because the overall prognosis with current standard of care, high dose chemotherapy and allogeneic transplantation, remains poor due to the emergence of refractory and relapsed disease. Here, we found expression of the transcription factor KLF4 in AML cell lines is not silenced through KLF4 gene methylation nor via proteasomal degradation. The deletion of KLF4 by CRISPR-CAS9 technology reduced cell growth and increased apoptosis in both NB4 and MonoMac-6 cell lines. Chemical induced differentiation of gene edited NB4 and MonoMac6 cells with ATRA and PMA respectively increased apoptosis and altered expression of differentiating markers CD11b and CD14. -
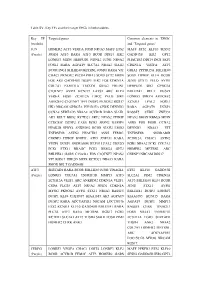
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -

ERG Promotes Transcriptional Reprogramming of Prostatic Epithelial Cells Toward a Cancer Stem Cell Phenotype
ERG promotes transcriptional reprogramming of prostatic epithelial cells toward a cancer stem cell phenotype Manju Sharma, Yubin Guo, Robert Bell, and Michael E. Cox. Vancouver Prostate Centre. Vancouver British Columbia, Canada Abstract Genomic rearrangements that result in aberrant expression of the proto-oncogene member of the erythroblast transformation-specific family of transcription factors, ERG, occur in almost half of prostate cancers. The most frequent rearrangements are the result of an interstitial deletion that results in fusion of androgen-responsive elements of serine 2-erythroblast transformation-specific-related gene with variable amino terminal ERG open reading frame exons. These TMPRSS2-ERG fusions are the most common cancer-associated mutation. They occur early in prostate cancer initiation, are associated with high Gleason score, and aggressive disease. When amplified and/or expressed at high level, TMPRSS2-ERG is associated with poor prognosis. Mirroring its functions in normal chondrocyte and endothelial cell differentiation, ERG expression in prostatic epithelial cells drives aberrant activation of transcriptional programs promoting migration, invasion, and epithelial-mesenchymal transition. Additionally, TMPRSS2-ERG status appears to enhance resistance to abiraterone and taxane therapies. We hypothesized that ERG expression can promote transformation of prostatic epithelial cells by directing transcriptional reprogramming towards a cancer stem cell phenotype. Transcriptional analysis of ERG-expressing RWPE1 prostatic epithelial cells (ERG-RWPE1) demonstrated up-regulation transcription of a panel of stem cell-related genes. When cultured under non- adherent conditions, ERG-RWPE1 cells selectively demonstrated the ability of form serially passable spheroids and dramatically increased expression of stem cell markers, PROM1, c-KIT and CD33. When re- plated in adherent conditions, cells re-established the expression pattern of parental adherent cultures. -

Expression of the Tumor Suppressor Krüppel-Like Factor 4 As a Prognostic Predictor for Colon Cancer
Published OnlineFirst August 10, 2010; DOI: 10.1158/1055-9965.EPI-10-0677 Cancer Research Article Epidemiology, Biomarkers & Prevention Expression of the Tumor Suppressor Krüppel-Like Factor 4 as a Prognostic Predictor for Colon Cancer Nilesh V. Patel1, Amr M. Ghaleb1, Mandayam O. Nandan1, and Vincent W. Yang1,2 Abstract Background: The zinc finger transcription factor Krüppel-like factor 4 (KLF4) regulates numerous physiologic processes, including proliferation, differentiation, and development. Studies also showed that KLF4 is involved in tumorigenesis and somatic cell reprogramming. Here, we aimed to assess whether KLF4 is a prognostic indicator for colon cancer. Methods: Levels of KLF4 were measured by immunohistochemical analysis of a tissue microarray contain- ing 367 independent colon cancer sections. Univariate data analysis was done in addition to construction of multivariate models with several clinicopathologic factors to evaluate KLF4 as an independent predictor of survival and cancer recurrence (disease-free survival). Results: Colon cancer tissues had significantly overall lower KLF4 levels compared with noncancer tissues (P < 0.0001). Using logistic regression, a trend was noted for decreased odds of KLF4 expression in higher stages of tumors. In univariate and multivariate analyses, KLF4 was a significant predictor of survival and recurrence. Conclusions: KLF4 expression is significantly downregulated in colon cancer, and loss of KLF4 is an independent predictor of survival and recurrence. Impact: These findings suggest that KLF4 may serve as a prognostic biomarker for colon cancer. Cancer Epidemiol Biomarkers Prev; 19(10); 2631–8. ©2010 AACR. Introduction markers independent of established clinicopathologic predictors of the disease. Despite substantial advances in the early diagnosis and Biomarker research in colorectal cancer is becoming treatment of colorectal cancer, it remains a disease with increasingly popular for a variety of clinical and research high morbidity and mortality.