Introduction of FRA Won't Contribute Towards the Harmonization Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PCB Annual Report 2018-19
Designed by PRESTIGE Annual Report 2018-2019 ANNUAL REPORT 2018-2019 Contents Foreword Men's domestic cricket Chairman's Report 1 Regional Inter-District 2018-2019 65 Managing Director's Report 4 Quaid-e-Azam Trophy 67 Overview of men's international cricket 5 Quaid-e-Azam Trophy Grade-II 69 Overview of women’s international/domestic cricket 7 One-Day Cup for Regions and Departments 71 Overview of men's domestic cricket 9 Quaid-e-Azam One-Day Cup 73 Overview of women’s game development 11 National T20 Cup 75 Overview of the Academies' programmes 13 HBL PSL 2019 77 Obituaries 16 Pakistan Cup 83 Patron's Trophy Grade-II 85 Men's international cricket (2018-2019) Women's domestic cricket Asia Cup 2018 19 Inter-Departmental T20 Women's Cricket Championship 89 Pakistan vs Australia in the UAE 21 PCB Triangular One-Day Women’s Cricket Tournament 2018-19 91 Pakistan vs New Zealand in the UAE 25 Pakistan in South Africa 27 Pathways cricket Pakistan in England 31 U13 Regional National T20 Tournament 95 U16 Regional National One-Day Tournament 97 Men's international cricket U16 Pentangular One-Day Tournament 99 (2017-2018) Inter-Region U19 Three-Day Tournament 101 Independence Cup 2018 Pakistan vs World XI 35 Inter-Region U19 One-Day Tournament 103 Pakistan vs Sri Lanka in the UAE and Lahore 37 Pentangular U19 T20 Cup 105 Pakistan in New Zealand 39 Pakistan A vs New Zealand A and England Lions in the UAE 106 West Indies in Karachi 41 Pakistan U16 vs Australia U16 in the UAE 109 Pakistan tour of Ireland, England and Scotland 43 Pakistan U16 in Bangladesh -

High-Throughput SNP Discovery and Genotyping for Constructing a Saturated Linkage Map of Chickpea (Cicer Arietinum L.)
DNA RESEARCH 19, 357–373, (2012) doi:10.1093/dnares/dss018 Advance Access publication on 3 August 2012 High-Throughput SNP Discovery and Genotyping for Constructing a Saturated Linkage Map of Chickpea (Cicer arietinum L.) RASHMI Gaur, SARWAR Azam, GANGA Jeena, AAMIR WASEEM Khan, SHALU Choudhary, MUKESH Jain, GITANJALI Yadav, AKHILESH K. Tyagi, DEBASIS Chattopadhyay, and SABHYATA Bhatia* National Institute of Plant Genome Research, Aruna Asaf Ali Marg, PO Box 10531, New Delhi 110067, India *To whom correspondence should be addressed. Tel. þ91 11-26735159. Fax. þ91 11-26741658. E-mail: [email protected] Edited by Satoshi Tabata (Received 18 April 2012; accepted 5 July 2012) Abstract The present study reports the large-scale discovery of genome-wide single-nucleotide polymorphisms (SNPs) in chickpea, identified mainly through the next generation sequencing of two genotypes, i.e. Cicer arietinum ICC4958 and its wild progenitor C. reticulatum PI489777, parents of an inter-specific reference mapping population of chickpea. Development and validation of a high-throughput SNP geno- typing assay based on Illumina’s GoldenGate Genotyping Technology and its application in building a high-resolution genetic linkage map of chickpea is described for the first time. In this study, 1022 SNPs were identified, of which 768 high-confidence SNPs were selected for designing the custom Oligo Pool All (CpOPA-I) for genotyping. Of these, 697 SNPs could be successfully used for genotyping, demonstrating a high success rate of 90.75%. Genotyping data of the 697 SNPs were compiled along with those of 368 co-dominant markers mapped in an earlier study, and a saturated genetic linkage map of chickpea was constructed. -

ICAB Monthly News Bulletin June 2021 Issue
June 2021 Number 377 icab news bulletin Monthly News Briefing from the Institute of Chartered Accountants of Bangladesh (ICAB) SME’s Contribution to GDP is Very Low in Comparison to Other Emerging Economies, Needs SMPs’ Support to Revive the Sector Speakers suggest at a Global Webinar SME’s Contribution to 1 GDP... Revive the Sector Auditing Software 3 Launched:.. audit begins President’s 4 Communication Inauguration of ICAB 7 Digital... Education Minister Some provisions of 8 Companies... Webinar Meeting with NBR 9 Chairman Meeting with CAG 9 Bangladesh Press Conference on 10 National Budget 2021 ICAB Team paid a 12 courtesy... Secretary, GoB Campus News 12 Chartered Accountancy- 13 A Career Choice Training on “Minimum 13 audit... each audit client” Virtual Members’ 14 Conference... forward’ Online Certificate Course 14 ... Audit (IS Audit)-5th Batch n Bangladesh SMEs are considered as situation has aggravated as cash-flow problem, Online Certificate Course 14 on... Based on ISA engine of economic growth that constitutes liquidity crisis, IT skill gap, reduction of Anwaruddin Chowdhury 15 I over 90 percent of business enterprises while production costs, shifting to e-commerce Participates... Meeting it is 97.60 percent in India, 99 percent in China, practices and digitalization of operations have Admission as Fellows 15 99.70 percent in Japan and 60 percent in emerged as new challenges to SME sector. Permission to Continue 15 Pakistan. SME contributes to GDP 20.25 Practice The Institute of Chartered Accountants of percent which is very low in comparison to other Permission to Start 16 Bangladesh (ICAB) organized the Global emerging economies. -

Evsjv‡`K †M‡RU
†iwR÷vW© bs wW G-1 evsjv‡`k †M‡RU AwZwi³ msL¨v KZ…©c¶ KZ…©K cÖKvwkZ iweevi, wW‡m¤^i 6, 2015 MYcÖRvZš¿x evsjv‡`k miKvi evsjv‡`k miKvix Kg© Kwgkb mwPevjq AvMviMuvI, †k‡ievsjv bMi, XvKv-1207| weÁwß ZvwiL : 06 wW‡m¤^i 2015 bs 80.110.038.00.00.001.2015(Ask-1)-171 AvM÷/2015 mv‡ji wmwbqi †¯‹‡j c‡`vbœwZ cixÿvq †h mKj Kg©KZ©v GK ev GKvwaK wel‡q/c‡Î DËxY© n‡q‡Qb Zuv‡`i bv‡gi ZvwjKv wb‡gœ cÖ`Ë nBj| Cadre Code & Name: ( 101 )-ACCOUNTING SL . NO REGISTRATION NO NAME & ID NO PAPERS 1 101001 MD. RABIUL AWAL ,00021976 1st,2nd & 3rd 2 101002 ZIAUR RAHMAN ,00018344 1st,2nd & 3rd 3 101003 MD. SOHEL RANA ,00022121 1st,2nd & 3rd 4 101004 MD. KHALED HASAN ,00018435 1st,2nd & 3rd 5 101006 JAHIRUL ISLAM ,018176 1st,2nd & 3rd 6 101007 MOHON KUMAR DAS ,00021675 2nd & 3rd 7 101008 MD. ANOWARUL HOQUE ,015178 1st,2nd & 3rd 8 101009 MD. GIAS UDDIN ,0018161 1st & 2nd 9 101010 MOHAMMAD KAMAL HOSSAIN ,017940 2nd 10 101012 BODIUL ALAM ,017981 2nd 11 101014 MOHAMMAD MAHFUZUR RAHMAN BHUIYAN, 00022565 1st,2nd & 3rd 12 101015 MADHUSUDAN MONDAL ,022059 1st,2nd & 3rd ( 10495 ) g~j¨ t UvKv 44.00 10496 evsjv‡`k †M‡RU, AwZwi³, wW‡m¤^i 6, 2015 SL . NO REGISTRATION NO NAME & ID NO PAPERS 13 101016 SHYAMAL CHAKROBORTY ,00022288 2nd & 3rd 14 101017 SWEETY RANI MODAK ,018376 3rd 15 101018 MD. SOHEL RANA ,00021891 1st,2nd & 3rd 16 101022 MD. SAIDUR RAHMAN ,021697 2nd 17 101026 MOHAMMAD SIDDIKUR RAHMAN , 00021700 2nd 18 101027 MAMUN KHAN ,017937 2nd 19 101029 MD. -

17Th Graduation Ceremony-2019 1 MILITARY INSTITUTE of SCIENCE and TECHNOLOGY (MIST) MIRPUR CANTONMENT
17th Graduation Ceremony-2019 1 MILITARY INSTITUTE OF SCIENCE AND TECHNOLOGY (MIST) MIRPUR CANTONMENT INAUGURAL CEREMONY OF MIST ON 19 APRIL 1998 HONOURABLE PRIME MINISTER OF PEOPLE’S REPUBLIC OF BANGLADESH SHEIKH HASINA UNVEILING THE FOUNDATION PLAQUE 1717thth Graduation Graduation Ceremony-2019 Ceremony-2019 22 MESSAGE Minister I am delighted to know that Military Institute of Science and Technology (MIST) is going to celebrate its 17th Graduation Ceremony on 19 March 2019. On this day, MIST will cross another milestone of success in the field of technical and knowledge based education. I firmly believe that establishment of MIST on 19th April 1998 by the Honorable Prime Minister Sheikh Hasina was a milestone in the history of technical education in Bangladesh towards materializing the dream of the Father of the Nation Bangabandhu Sheikh Mujibur Rahman to build “Sonar Bangla”. MIST is contributing significantly in building technology and knowledge based prosperous nation. On this day, I would like to congratulate all graduates and wish them all the very best in pursuing their career in future. I would like to convey the message to all graduates that you have worked hard, and your parents, families, faculty members and the institute itself have sacrificed much to get you here, to become a proud engineer. Make them and your institute proud through your contributions, especially by being hard working, innovative, committed and ethical in your profession. I am glad to know that within a short span of only 19 years. MIST has grown into a well-rounded institute with its own campus and 12 departments under four faculties where more than two thousand five hundred civil and military students pursue their study. -

Annual Report Have Been Captured in the Campus of NIAB Annual Report | 2019-20
Annual NIAB Report 2019-20 Photographs used in the Annual Report have been captured in the campus of NIAB Annual Report | 2019-20 1 2 Annual Report | 2019-20 S. No. TABLE OF CONTENTS Page No. 1. Mission and Vision of NIAB 05 2. From the desk of Director 06 3. Research Projects: 09 A. Animal Genomics and Reproduction 09 a. Livestock Genomics for Cattle Improvement and Transgenic Farmed Animals (Dr. 10 Subeer S Majumdar) b. Reproductive Biology, Gametogenesis, Oocyte atresia, DNA damage response and 15 repair pathways (Dr. H.B.D. Prasada Rao) c. Biopharming Using Farmed Animals and Avenues for Obtaining Sperm with Elite 19 Trait (Dr. Nirmalya Ganguli) d. Aptamer and antibody based point-of-care diagnostics for better animal production 22 and health ( Dr. Pankaj Suman) B. Animal Health 26 a. Microbial Patho biology and One Health (Dr. Nagendra R. Hegde) 27 b. Understanding the virulence mechanisms of the zoonotic pathogen, Brucella and 33 development of improved vaccines and diagnostic assays for animal and human brucellosis (Dr. Girish K Radhakrishnan) c. Development of Leptospirosis vaccine and novel veterinary adjuvants (Dr. Syed M 36 Faisal) d. Host Pathogen Interaction Studies on Animal and Avian Viruses (Dr. Madhuri 39 Subbiah) e. Host pathogen interaction studies on animal parasites (Dr. Anand Srivastava) 42 f. Study of Virulence, Antimicrobial Resistance and Host Pathogenesis during 47 Intracellular Pathogens infection (Dr. Paresh Sharma) g. Role of CDK-related kinases (Crks) in transcription regulation in Toxoplasma gondii 51 (Dr. Abhijit S Deshmukh) h. Tuberculosis and other zoonotic diseases of livestock: Molecular pathogenesis and 55 Intervention Strategies (Dr. -
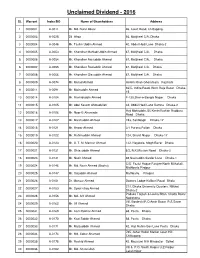
Unclaimed Divident
Unclaimed Dividend - 2016 SL Warrant Index/BO Name of Shareholders Address 1 0000001 A-0011 Mr. Md. Nurul Absar 46, Court Road, Chittagong. 2 0000003 A-0025 Dil Afroz 66, Motijheel C/A, Dhaka 3 0000004 A-0048 Mr. Taslim Uddin Ahmed 40, Abdul Hadi Lane Dhaka-2 4 0000005 A-0053 Mr. Khondker Mahtab Uddin Ahmed 67, Motijheel C/A. Dhaka 5 0000006 A-0054 Mr. Khondker Raisuddin Ahmed 67, Motijheel C/A. Dhaka 6 0000007 A-0055 Mr. Khondker Raziuddin Ahmed 67, Motijheel C/A. Dhaka 7 0000008 A-0056 Mr. Khondker Giasuddin Ahmed 67, Motijheel C/A. Dhaka 8 0000009 A-0074 Mr. Murad Ahmed Hetem Khan Ghoramara Rajshahi 66/C, Indira Road, West Raja Bazar Dhaka- 9 0000011 A-0091 Mr. Moinuddin Ahmed 15 10 0000014 A-0104 Mr. Rashiduddin Ahmed F-130,Sher-e-Bangla Nagar Dhaka 11 0000015 A-0105 Mr. Abul Kasem Ahmadullah 34, Abdul Hadi Lane Ramna Dhaka-2 Haji Mohiuddin, 56 KiminiVushan Rudduru 12 0000016 A-0106 Mr. Noor-E-Ahameda Road Dhaka 13 0000017 A-0107 Mr. Masihuddin Ahmed 193, Santibagh Dhaka-17 14 0000018 A-0121 Mr. Anwar Ahmed 3/1 Purana Paltan Dhaka 15 0000019 A-0122 Mr. Rahimuddin Ahmed 124, Shanti Nagar Dhaka-17 16 0000020 A-0123 Mr. A. T. M. Mansur Ahmed 132, Nayatola, Mogh Bazar Dhaka 17 0000021 A-0131 Mr. Ghiasuddin Ahmed 8/2, R.K.Mission Road Dhaka-3 18 0000023 A-0141 Mr. Nazir Ahmed 48,Nasiruddin Sardar Lane Dhaka-1 C/O. Fazlul Haque Farajee North Mithakali, 19 0000024 A-0146 Mr. -

ICAB Monthly News Bulletin July 2020 Issue
Number 366 July 2020 ICAB NEWS BULLETIN Monthly News Briefing from the Institute of Chartered Accountants of Bangladesh (ICAB) Strongest and Resilient Economies Need Trustworthy Information Strongest and Resilient 1 Economies... Information CRC Donates Medical 3 Equipment... COVID-19 President’s Communication 4 Trustworthy Financial 6 Reporting... Conference Virtual SAFA Forum on 7 Challenges... Outbreak Recommendations for 7 Monetary Policy Virtual Members’ 8 Conference... Challenges Sabbir Ahmed Participated 9 in ICMA... Webinar he strongest and most resilient Salman F Rahman MP, Private Sector Industry Admission as Fellows 9 economies will be those where people and Investment Adviser to the Honorable Prime T can trust information they receive; the Minister of the Government of the People’s Permission to Start 9 leaders can make well-informed decisions, Republic of Bangladesh attended the Practice businesses can be held to account and public conference as the Chief Guest. Manil Permission to Continue 9 finances are transparent. The skills that the Jaysinghe FCA, President CA Sri Lanka was Practice accountancy profession instills in measuring, present as Special Guest while Dr. In-Ki Joo, reporting and assuring –will be fundamental in President of the International Federation of Permission to Join as 9 Accountants (IFAC) was present as Guest of Partner building a World of Strong Economies. The technical skills, ethical approach and the Honour. Robert Hodgkinson FCA, Executive Permission to Open CA 10 integrity are very important for the purpose. Director, Technical at the Institute of Chartered Firm Under the current pressures of the pandemic, Accountants in England and Wales (ICAEW), Permission to train Articled 10 everybody CAs, in particular should take UK presented the keynote paper. -

5. Chronological List of Members.Indd
Chronological List of Members SL No. Enrl. No. Name of Members 1 9 Md Jainul Abedin FCA 2 16 Md Muzaffar Ahmed, FCA 3 24 Kamal Ziaul Islam FCA 4 28 Hari Sadhan Dhar FCA 5 30 A K M Rafi qul Islam FCA 6 37 AKM Mosharraf Hossain FCA 7 38 Kazi Aminul Huque FCA 8 39 Nazrul Islam FCA 9 41 Abdul Hafi z Choudhury FCA 10 43 Beg Mohammed Nurul Azim FCA 11 45 Muhammad Abdus Sattar FCA 12 46 ABM Azizuddin FCA 13 48 Abdul Wahab FCA 14 49 Md Obaidur Rahman FCA 15 51 Abdur Rouf Bhuiya FCA 16 53 Anil Chandra Nath FCA 17 56 Amanullah Khan FCA 18 57 Badrul Alam FCA 19 60 Abu Nasar Altaf Hussain Siddiqui FCA 20 61 Md Abu Taleb Talukder FCA 21 63 Syed Fazlul Haque FCA 22 64 Md Abdul Majid FCA 23 66 ASM Ataul Karim FCA 24 68 Kazi Zahirul Kabir FCA 25 69 M Ashraful Haque FCA 26 73 M A Quadir Mollah FCA 27 74 Abul Kashem FCA 28 78 Md Mushtaque Ahmed FCA 29 80 Mainuddin Ahmed FCA 30 84 Md Azharul Islam FCA 31 89 Shahid Uddin Ahmed FCA 32 92 Sultan Ahmed FCA 33 94 Md Abu Taher FCA 34 95 M Idris Ali FCA 35 97 Muhammed Abdul Halim Gaznavi FCA 36 101 Abu Khaled Mohammad Akhtaruzzaman FCA 37 103 Md Zahirul Islam FCA 38 104 AKM Golam Mostafa FCA 39 105 Howlader Mahfel Huq FCA 40 107 AHM Mustafa Kamal FCA 41 110 Sheikh Abdul Hafi z FCA 42 111 Mohammed Salahuddin FCA 502 MEMBERS AND FIRMS 2019-2020 SL No. -

2013 ANNUAL REPORT I Am a Little Obsessed with Fertilizer
2013 ANNUAL REPORT I am a little obsessed with fertilizer. I mean I’m fascinated with its role, “not with using it. Two out of every five people on Earth today owe their lives to the higher crop outputs that fertilizer has made possible. It helped fuel the Green Revolution, an explosion of agricultural productivity that lifted hundreds of millions of people around the world out of poverty.” – Bill Gates, “Here’s My Plan to Improve Our World and How You Can Help,” November 12, 2013 CONTENTS 2 IFDC Around the World 4 2014 IFDC Board of Directors Message from the Chairman of the Board and the 5 President and Chief Executive Officer 40 Years: Amidst Evolution, Focus on Smallholder 6 Farmers Remains 10 Highlights 12 Annual Activities Align With Strategic Plan 16 2013 Strategy Workshops 20 2013 Special Initiatives 22 EurAsia Division 26 East and Southern Africa Division 32 North and West Africa Division 38 Office of Programs 42 Training & Workshop Coordination Unit 44 2013 Selected Articles, Presentations and Studies 46 IFDC Offices and Staff 57 Revenue Sources 58 Financial Highlights 60 Acronyms and Abbreviations and Publication Credits ANNUAL REPORT 2013 | 1 Countries With IFDC Projects in 2013 Bangladesh Benin Burkina Faso Burundi Cape Verde Chad Côte d’Ivoire Democratic Republic of Congo Ethiopia Gambia Ghana Guinea Guinea-Bissau Kenya Kyrgyzstan Liberia Mali Mozambique Niger Nigeria Rwanda Senegal Sierra Leone South Sudan Tajikistan Tanzania Togo Uganda United States of America IFDC Zambia Zimbabwe Around the World 2 | 2013 ANNUAL REPORT IFDC Around the World For a complete list of countries in which IFDC has worked, visit www.ifdc.org/About/Map_IFDC_in_the_World. -

1 Sl No. Student Name Reg. No. Roll No. 1 Md. Mayeen Uddin 2016100848 DHK-29720
The Institute of Cost and Management Accountants of Bangladesh CMA June - 2020 EXAMINATION RESULTS (New ) FINAL COMPLETE(New) Number of Students: 1 Sl No. Student Name Reg. No. Roll No. 1 Md. Mayeen Uddin 2016100848 DHK-29720 Page 1 of 1 The Institute of Cost and Management Accountants of Bangladesh CMA JUNE-2020 EXAMINATION RESULTS FINAL COMPLETE (Old Syllabus) Number of Students: 52 Sl No. Student Name Reg. No. Roll No. 1 BIPLOB KUMER SANNYASI 20114113 DHK-29484 2 MD. ABU BAKAR SIDDIQUE 2018101201 DHK-29510 3 NAVID AHSAN 20121998 DHK-29533 4 MD. ABDUR RAHIM 20113450 DHK-29578 5 MOHAMMAD HASAN AKHTER 2020101077 DHK-29654 6 MD. ALAUDDIN 2017200968 DHK-29656 7 MD. SHAKAWATH HOSSAIN BHUYAN 20114111 DHK-29763 8 MAHAMMUD KAYSAR 2019201248 DHK-29767 9 MUHAMMAD SHAMEEM MAHFOOZ 2017201336 DHK-29841 10 MOHAMMAD ASHRAFUR RAHMAN 2020200697 DHK-29945 11 ASHRAFUL ALAM 20121572 DHK-29947 12 ALAUL HASAN BHUIYAN D-20143246 DHK-29950 13 MD. MERAZ HOSEN 20143192 DHK-29955 14 TANIA YASMIN KHAN 2018200696 DHK-29965 15 SAGAR KUMAR KUNDU 2020101138 DHK-29973 16 SK. MD. MOSHARRAF HOSSAIN 2018101500 DHK-30023 17 KAZI MOHAMMAD MOINUL ISLAM 2017201234 DHK-30070 18 MD. MOSHAHEB KAKKA 2017201317 DHK-30079 19 A. MOMIN 20111064 DHK-30088 20 MOHAMMAD SALAHUDDIN 20153952 DHK-30107 21 Mohammad Rafiqul Islam 2016101755 DHK-30123 22 SAIFUL ISLAM 2019100975 DHK-30129 23 MD. NURUL ISLAM MOLLA 20122179 DHK-30154 24 MOHAMMED ALAMGIR HOSSAIN 2020200685 DHK-30269 25 ZABAD MOHAMMAD MUSAVI 2019201219 DHK-30342 26 MD. ARIFUL ISLAM BHUIYAN 20131771 DHK-30451 27 MD. IMAM HOSSAIN MOZUMDER 20123388 DHK-30477 28 WAHID SABER KANCHAN 20141745 DHK-30492 29 MD. -

Fazlul Huq and Bengal Politics in the Years Before the Second Partition of Bengal (1947)
6 Fazlul Huq and Bengal Politics in the years before the Second Partition of Bengal (1947) On 24 April 1943, H.E. the Governor Sir John Herbert with the concurrence of the Viceroy, issued a proclamation by which he revoked the provision of Section 93 and invited the leader of the Muslim League Assembly Party, Khwaja Sir Nazimuddin to form a new ministry. The aspirant Nazimuddin who made constant effort to collect support in order to form a new ministry, ultimately managed to have the support of 140 members in the Bengal Assembly (consisting of 250 membrs). On the other hand, the Opposition (led by Fazlul Huq) had 108 members. Following the dismissal of the Second Huq Ministry, a large number of Muslim M.L.A.s thought it expedient to cross the floor and join the League because of Governor‟s inclination towards Nazimuddin whom he invited to form a new ministry. As they flocked towards the League, the strength of the League Parliamentary Party rose from 40 to 79. Abul Mansur Ahmed, an eminent journalist of those times, narrated this trend in Muslim politics in his own fashion that „everyone had formed the impression that the political character of the Muslim Legislators was such that whoever formed a ministry the majority would join him‟1 which could be applicable to many of the Hindu members of the House as well. Nazimuddin got full support of the European Group consisting of 25 members in the Assembly. Besides, he had been able to win the confidence of the Anglo-Indians and a section of the Scheduled Caste and a few other members of the Assembly.