NZFSS Newsletter 51 (2012)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Detailed Itinerary [ID: 1136]](https://docslib.b-cdn.net/cover/9604/detailed-itinerary-id-1136-239604.webp)
Detailed Itinerary [ID: 1136]
Daniel Collins - Fine Travel 64 9 363 2754 www.finetravel.co.nz Land of the Rings A tour appealing to those Frodo and Gandalf fans, with visits to official sites used in the filming of The Lord of the Rings and The Hobbit. Starts in: Auckland Finishes in: Queenstown Length: 9days / 8nights Accommodation: Hotel 3 star Can be customised: Yes This itinerary can be customised to suit you perfectly. We can add more days, remove days, change accommodations, mix it up, add activities to suit your interests or simply design and create something from scratch. Call us today to get your custom New Zealand itinerary underway. Inclusions: Includes: All coach transport Includes: All pick ups/drop offs at destinations Includes: Comprehensive tour pack (detailed Includes: 24/7 support while touring in New itinerary, driving, instructions, map/guidebooks, Zealand brochures) Included activity: Auckland to Rotorua including Included activity: Private transfer Christchurch Waitomo and Hobbiton with GreatSights Airport to your accommodation Included activity: Air New Zealand flight Rotorua Included activity: Rotorua Accommodation to to Christchurch Rotorua Airport - Private Transfer Included activity: Rotorua Duck Tours City & Included activity: Lord of the Rings Edoras Tour Lakes Tour Included activity: Christchurch to Queenstown Included activity: One Day in Middle Earth Full via Mount Cook with GreatSights Day Tour Included activity: Safari of the Scenes Wakatipu Included activity: Queenstown to Te Anau with Basin Tour GreatSights Included activity: Pure Wilderness Jet Boat Included activity: Milford Sound Nature Cruise : Meals included: 1 lunch, 1 dinner For a detailed copy of this itinerary go to http://finetravel.nzwt.co.nz/tour.php?tour_id=1136 or call us on 64 9 363 2754 Day 1 Auckland to Rotorua including Waitomo and Hobbiton Experience with GreatSights Take a journey into the heart of the Central North Island, taking in the Waitomo Caves and Hobbiton Movie Set on a full-day sightseeing tour. -
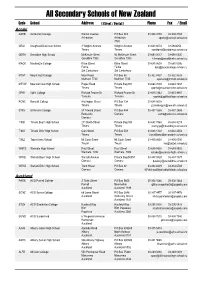
Secondary Schools of New Zealand
All Secondary Schools of New Zealand Code School Address ( Street / Postal ) Phone Fax / Email Aoraki ASHB Ashburton College Walnut Avenue PO Box 204 03-308 4193 03-308 2104 Ashburton Ashburton [email protected] 7740 CRAI Craighead Diocesan School 3 Wrights Avenue Wrights Avenue 03-688 6074 03 6842250 Timaru Timaru [email protected] GERA Geraldine High School McKenzie Street 93 McKenzie Street 03-693 0017 03-693 0020 Geraldine 7930 Geraldine 7930 [email protected] MACK Mackenzie College Kirke Street Kirke Street 03-685 8603 03 685 8296 Fairlie Fairlie [email protected] Sth Canterbury Sth Canterbury MTHT Mount Hutt College Main Road PO Box 58 03-302 8437 03-302 8328 Methven 7730 Methven 7745 [email protected] MTVW Mountainview High School Pages Road Private Bag 907 03-684 7039 03-684 7037 Timaru Timaru [email protected] OPHI Opihi College Richard Pearse Dr Richard Pearse Dr 03-615 7442 03-615 9987 Temuka Temuka [email protected] RONC Roncalli College Wellington Street PO Box 138 03-688 6003 Timaru Timaru [email protected] STKV St Kevin's College 57 Taward Street PO Box 444 03-437 1665 03-437 2469 Redcastle Oamaru [email protected] Oamaru TIMB Timaru Boys' High School 211 North Street Private Bag 903 03-687 7560 03-688 8219 Timaru Timaru [email protected] TIMG Timaru Girls' High School Cain Street PO Box 558 03-688 1122 03-688 4254 Timaru Timaru [email protected] TWIZ Twizel Area School Mt Cook Street Mt Cook Street -

Cyanosed: a Workshop on Benthic Cyanobacteria and Cyanotoxins
AGENDA ITEM 3 OPEN DISCUSSION & ANNOUNCEMENTS OUTLINE OF TOPICS • CyanoSED: A Workshop on Benthic Cyanobacteria and Cyanotoxins – Overview and Objectives • Special Issue in the Open Access Journal TOXINS • 1st Cyanobacteria Twitter Conference • Journal Publications • Upcoming HAB Meetings • EPA FHAB Newsletter • 2019 Benthic HABs Presentations – Call for Presenters UNCLASSIFIED File Name 3 CyanoSED: A Workshop on Benthic Cyanobacteria and Cyanotoxins Overview and Objectives Kaytee Pokrzywinski, Tim Davis, Susie Wood, Jim Lazorchak, Brooke Stevens, Jonathan Puddick, Andrew McQueen, Karen Keil, Mike Habberfield USACE-ERDC, BGSU, Cawthron Institute, US EPA and USACE-LRB US Army Corps of Engineers • Engineer Research and Development Center DISCOVER | DEVELOP | DELIVER UNCLASSIFIED UNCLASSIFIED 4 Purpose and Inspiration • The primary purpose of the workshop is to identify knowledge gaps and prioritize research needs on issues surrounding ‘benthic cyanobacteria’. • Two ERDC reports funded by our Dredging Operational Technical Support Program (DOTS) US Army Corps of Engineers • Engineer Research and Development Center UNCLASSIFIED UNCLASSIFIED 5 Context • What do we mean by benthic cyanobacteria? • In, on or near sediment or other ‘substrate’ • Planktonic vs periphytic (mats, films) • Filamentous vs single celled • Toxic vs nuisance Quiblier et al. 2013 Water Research • Lakes vs rivers • Benthic-pelagic coupling CyanoSED Cyanobacteria + Sediment… in any conformation or condition Photo: Jeff Sowards, UFL Hense & Beckman 2006 Ecological modeling -

Clifden Suspension Bridge, Waiau River
th IPENZ Engineering Heritage Register Report Clifden Suspension Bridge, Waiau River Written by: Karen Astwood Date: 3 September 2012 Clifden Suspension Bridge, newly completed, circa February 1899. Collection of Southland Museum and Art Gallery 1 Contents A. General information ........................................................................................................... 3 B. Description ......................................................................................................................... 5 Summary ................................................................................................................................. 5 Historical narrative .................................................................................................................... 6 Social narrative ...................................................................................................................... 11 Physical narrative ................................................................................................................... 12 C. Assessment of significance ............................................................................................. 16 D. Supporting information ...................................................................................................... 17 List of supporting documents ................................................................................................... 17 Bibliography .......................................................................................................................... -

PCR), Quantitative PCR and Metabarcoding for Species‐Specific Detection in Environmental DNA
Received: 18 January 2019 | Revised: 7 June 2019 | Accepted: 10 June 2019 DOI: 10.1111/1755‐0998.13055 RESOURCE ARTICLE A comparison of droplet digital polymerase chain reaction (PCR), quantitative PCR and metabarcoding for species‐specific detection in environmental DNA Susanna A. Wood1 | Xavier Pochon1,2 | Olivier Laroche1,3 | Ulla von Ammon1,2 | Janet Adamson1 | Anastasija Zaiko1,2 1Coastal and Freshwater Group, Cawthron Institute, Nelson, New Zealand Abstract 2Institute of Marine Science, University of Targeted species‐specific and community‐wide molecular diagnostics tools are Auckland, Auckland, New Zealand being used with increasing frequency to detect invasive or rare species. Few studies 3Department of Oceanography, School of Ocean and Earth Science and have compared the sensitivity and specificity of these approaches. In the present Technology, University of Hawaii at Manoa, study environmental DNA from 90 filtered seawater and 120 biofouling samples Honolulu, HI, USA was analyzed with quantitative PCR (qPCR), droplet digital PCR (ddPCR) and meta‐ Correspondence barcoding targeting the cytochrome c oxidase I (COI) and 18S rRNA genes for the Susanna A. Wood, Coastal and Freshwater Group, Cawthron Institute, Nelson, New Mediterranean fanworm Sabella spallanzanii. The qPCR analyses detected S. spallan‐ Zealand. zanii in 53% of water and 85% of biofouling samples. Using ddPCR S. spallanzanii was Email: [email protected] detected in 61% of water of water and 95% of biofouling samples. There were strong Funding information relationships between COI copy numbers determined via qPCR and ddPCR (water New Zealand Government's Strategic 2 2 Science Investment Fund (SSIF) through R = 0.81, p < .001, biofouling R = 0.68, p < .001); however, qPCR copy numbers were the NIWA Coasts and Oceans Research on average 125‐fold lower than those measured using ddPCR. -

Lake Tekapo to Twizel Highlights
AORAKI/MT COOK WHITE HORSE HILL CAMPGROUND MOUNT COOK VILLAGE BURNETT MOUNTAINS MOUNT COOK AIRPORT TASMAN POINT Tasman Valley Track FRED’S STREAM TASMAN RIVER JOLLIE RIVER SH80 Jollie Carpark Braemar-Mount Cook Station Rd 800 TEKAPO TWIZEL 700 54km ALTERNATIVEGLENTANNER PARK CENTRE ROUTE: Lake Tekapo to Twizel 600 LANDSLIP CREEK ELEVATION Fitness: Easy • Skill: Easy • Traffic: Low • Grade: 2 500 400 KM LAKE PUKAKI 0 10 20 30 40 50 MT JOHN OBSERVATORY LAKE TEKAPO BRAEMAR ROAD Tekapo Powerhouse Rd LAKE TEKAPO TEKAPO A POWER STATION SH8 3km TRAIL GUARDIAN Hayman Rd SALMON FARM TO SALMON SHOP Tekapo Canal Rd PATTERSONS PONDS 9km TEKAPO CANAL 15km Tekapo Canal Rd LAKE PUKAKI SALMON FARM 24km TEKAPO RIVER TEKAPO B POWER STATION Hayman Road LAKE TEKAPO 30km Lakeside Dr Te kapo-Twizel Rd Church of the 8 Good Shepherd Dog Monument MARY RANGES SH80 35km r s D TEKAPO RIVERe SH8 r r 44km C e i e Pi g n on n Roto Pl o i e a e P SALMON SHOP r r D o r A Scott Pond Aorangi Cres 8 PUKAKI CANAL SH8 F Rd airlie-Tekapo PUKAKI RIVER Allan St Glen Lyon Rd Glen Lyon Rd LAKE TEKAPO Andrew Don Dr Old Glen Lyon Rd Pukaki Flats Track Murray Pl TWIZEL PUKAKI FLATS Mapwww.alps2ocean.com current as of 28/7/17 N 54km OHAU CANAL LAKE RUATANIWHA 0 1 2 3 4 5km KEY: Onroad Off-road trail SH8 Scale The alternative route begins in the at the Mt Cook Alpine Salmon shop 44km . You then cross the Tekapo township near the police highway and follow the trail across Pukaki Flats – an expansive Highlights: station. -

Middle Earth: Hobbit & Lord of the Rings Tour
MIDDLE EARTH: HOBBIT & LORD OF THE RINGS TOUR 16 DAY MIDDLE EARTH: HOBBIT & LORD OF THE RINGS TOUR YOUR LOGO PRICE ON 16 DAYS MIDDLE EARTH: HOBBIT & LORD OF THE RINGS TOUR REQUEST Day 1 ARRIVE AUCKLAND Day 5 OHAKUNE / WELLINGTON Welcome to New Zealand! We are met on arrival at Auckland This morning we drive to the Mangawhero Falls and the river bed where International Airport before being transferred to our hotel. Tonight, a Smeagol chased and caught a fish, before heading south again across the welcome dinner is served at the hotel. Central Plateau and through the Manawatu Gorge to arrive at the garden of Fernside, the location of Lothlorién in Featherston. Continue south Day 2 AUCKLAND / WAITOMO CAVES / HOBBITON / ROTORUA before arriving into New Zealand’s capital city Wellington, home to many We depart Auckland and travel south crossing the Bombay Hills through the of the LOTR actors and crew during production. dairy rich Waikato countryside to the famous Waitomo Caves. Here we take a guided tour through the amazing limestone caves and into the magical Day 6 WELLINGTON Glowworm Grotto – lit by millions of glow-worms. From Waitomo we travel In central Wellington we walk to the summit of Mt Victoria (Outer Shire) to Matamata to experience the real Middle-Earth with a visit to the Hobbiton and visit the Embassy Theatre – home to the Australasian premieres of Movie Set. During the tour, our guides escorts us through the ten-acre site ‘The Fellowship of the Ring’ and ‘The Two Towers’ and world premiere recounting fascinating details of how the Hobbiton set was created. -

Enhancing the Conservation Value of the Birchwood Wetland
Enhancing the Conservation Value of the Birchwood Wetland Robyn Newham and Courtney Quirin A report submitted in partial fulfilment of the Post-graduate Diploma in Wildlife Management University of Otago Year 2007 University of Otago Department of Zoology P.O. Box 56, Dunedin New Zealand WLM Report Number: 203 Enhancing the Conservation Value of the Birchwood Wetland Robyn Newham Courtney Quirin 1 EXECUTIVE SUMMARY The Birchwood wetland area has recently been included within the Ahuriri Conservation Park in the MacKenzie Basin, South Island, New Zealand. The wetland area is ranked as an ‘outstanding’ site of wildlife interest, providing breeding grounds for black stilt, black-fronted tern, large wrybill and Australian bittern populations, in addition to providing essential habitat for other waterfowl. However, past anthropogenic activities, such as drainage, recreation and introduced species, have degraded the Birchwood wetland. The aims of this research and management proposal attempt to increase the productivity of the wetland system, which serves to expand and improve the suitability of habitat for target birds and to enhance the awareness of the public through conservation education. Our research and management plan draws on past wetland enhancement strategies, in addition to incorporating site specific data and original thought. Recommendations A combined ecological and social approach to the management of the Birchwood wetland was proposed. ECOLOGICAL o Due to the complete lack of information about the ecological processes within the wetland, intensive research of the hydrology, geology, invertebrate and vegetation composition was recommended. o Research is critical as modification to the landscape and system may have serious long-term consequences to the Birchwood wetland and neighboring ecological systems. -

I-SITE Visitor Information Centres
www.isite.nz FIND YOUR NEW THING AT i-SITE Get help from i-SITE local experts. Live chat, free phone or in-person at over 60 locations. Redwoods Treewalk, Rotorua tairawhitigisborne.co.nz NORTHLAND THE COROMANDEL / LAKE TAUPŌ/ 42 Palmerston North i-SITE WEST COAST CENTRAL OTAGO/ BAY OF PLENTY RUAPEHU The Square, PALMERSTON NORTH SOUTHERN LAKES northlandnz.com (06) 350 1922 For the latest westcoastnz.com Cape Reinga/ information, including lakewanaka.co.nz thecoromandel.com lovetaupo.com Tararua i-SITE Te Rerenga Wairua Far North i-SITE (Kaitaia) 43 live chat visit 56 Westport i-SITE queenstownnz.co.nz 1 bayofplentynz.com visitruapehu.com 45 Vogel Street, WOODVILLE Te Ahu, Cnr Matthews Ave & Coal Town Museum, fiordland.org.nz rotoruanz.com (06) 376 0217 123 Palmerston Street South Street, KAITAIA isite.nz centralotagonz.com 31 Taupō i-SITE WESTPORT | (03) 789 6658 Maungataniwha (09) 408 9450 Whitianga i-SITE Foxton i-SITE Kaitaia Forest Bay of Islands 44 Herekino Omahuta 16 Raetea Forest Kerikeri or free phone 30 Tongariro Street, TAUPŌ Forest Forest Puketi Forest Opua Waikino 66 Albert Street, WHITIANGA Cnr Main & Wharf Streets, Forest Forest Warawara Poor Knights Islands (07) 376 0027 Forest Kaikohe Russell Hokianga i-SITE Forest Marine Reserve 0800 474 830 DOC Paparoa National 2 Kaiikanui Twin Coast FOXTON | (06) 366 0999 Forest (07) 866 5555 Cycle Trail Mataraua 57 Forest Waipoua Park Visitor Centre DOC Tititea/Mt Aspiring 29 State Highway 12, OPONONI, Forest Marlborough WHANGAREI 69 Taumarunui i-SITE Forest Pukenui Forest -

THE NEW ZEALAND GAZETTE 208I7
5 OCTOBER THE NEW ZEALAND GAZETTE 208i7 Waitepipi Stream Tributary of the Mangapu Stream. Map Boddington Range .. Mountain range lying between the Alma reference, approximatelyN. 82/5377. and Acheron Rivers and extending gener Waiteti .. For the locality situated approximately 4 ally north-eastwards from Mount North miles south of Te Kuiti. Also for stream umberland to Trig. D. and viaduct in vicinity. Boundary Stream .. Tributary of the Waiau River. Map re Weraroa .. For Trig. Station in Block XII, Wairoa ference, S. 54/0071. Instead of "Deep Survey District. Map reference, N. 137/ Stream". 293023. Bradley, Mount For feature, height 2,805 ft, in the Banks Whakairi Heights .. For flat topped mountain in the upper Peninsula hills system. Map reference, Kauaeranga River area, Thames County. S. 84/0636. Instead of "Mount Herbert". Map reference, N. 49/156374. Instead of Cow Stream Tributary of Waiau River. Map reference, "Table Mountain". S. 54/9870. Instead of "William Stream". Culverden Range Mountain range commencing at Mount TARANAKI LAND DISTRICT Culverden and extending generally west Name Situation and Remarks erly and northerly to "Pahau Pass". Doubtful Range Mountain range extending generally south Cataract Stream For the tributary of Stony River in Egmont easterly from a point south of Amuri Pass National Park, approximately 30 chains on the main divide to Trig. Z. and lying east of Paul Stream. south of the Doubtful River. The Cataracts For the waterfalls on "Cataract Stream". Glynn Wye Range .. Mountain range commencing at Trig. V, WELLINGTON LAND DISTRICT Mount Longfellow, and extending gener ally easterly to Trig. S, La Grippe, on the Name Situation and Remarks Organ Range. -

SIRG Ruapehu
Snow and Ice Research Group (SIRG) New Zealand Annual Workshop Mt Ruapehu 12th to 14th February 2007 Ruapehu Crater Lake from Tahurangi (2797m). Photo by Andrew Mackintosh PRESENTATION TIMETABLE Time Presenter Topic Monday 2:00 Heather and Welcome & Introduction Andrew 2:15 Trevor Chinn Glacier Mass Balance Measurements in N.Z. History and Development 2:30 Jordy Hendrix (for Glacier volume changes in the Southern Alps - Trends and Jim Salinger) variations from snowline monitoring 2:45 Andrew On the response of maritime glaciers to climatic change Mackintosh 3:00 Afternoon tea 3:30 Dorothea Stumm Index-stake Mass-balance measurements on Rolleston Glacier 3:45 Pascal Sirguey Operational and Improved Snow Mapping at Subpixel resolution in the Waitaki Basin using MODIS/TERRA 4:00 Tim Kerr Comparison of CROSS-Mountain precipitation profiles from around the world 4:15 Brian Anderson Response of KA ROIMATA O HINE HUKATERE Franz Josef Glacier to Climate Change 4:30 Uwe Morgenstern Ice dating on Tasman, Franz-Josef and Fox Glaciers 4:45 Simon Allen Geomorphic Hazard Modelling in the Mount Cook Region: Initial Results from ASTER based Glacial Lake MAPPING 5:00 Wolfgang Rack Tidal flexure and ice flux in the grounding zone of Jutulstraumen, Antarctica 7.00 pm Dinner at Lodge 8.00 pm Harry Keys After dinner talk about Mt Ruapehu Tuesday or Wednesday depending on weather 8:30 Shelly McDonnell Drainage system development of the Wright Lower Glacier, Antarctica, over a season 8:45 Martin Brook Late Quaternary glacial history of Mt Allen, southern Stewart -
Fiordland Day Walks Te Wāhipounamu – South West New Zealand World Heritage Area
FIORDLAND SOUTHLAND Fiordland Day Walks Te Wāhipounamu – South West New Zealand World Heritage Area South West New Zealand is one of the great wilderness areas of the Southern Hemisphere. Known to Māori as Te Wāhipounamu (the place of greenstone), the South West New Zealand World Heritage Area incorporates Aoraki/Mount Cook, Westland Tai Poutini, Fiordland and Mount Aspiring national parks, covering 2.6 million hectares. World Heritage is a global concept that identifies natural and cultural sites of world significance, places so special that protecting them is of concern for all people. Some of the best examples of animals and plants once found on the ancient supercontinent Gondwana live in the World Heritage Area. Left: Lake Marian in Fiordland National Park. Photo: Henryk Welle Contents Fiordland National Park 3 Be prepared 4 History 5 Weather 6 Natural history 6 Formation ������������������������������������������������������� 7 Fiordland’s special birds 8 Marine life 10 Dogs and other pets 10 Te Rua-o-te-moko/Fiordland National Park Visitor Centre 11 Avalanches 11 Walks from the Milford Road Highway ����������������������������� 13 Walking tracks around Te Anau ����������� 21 Punanga Manu o Te Anau/ Te Anau Bird Sanctuary 28 Walks around Manapouri 31 Walking tracks around Monowai Lake, Borland and the Grebe valley ��������������� 37 Walking tracks around Lake Hauroko and the south coast 41 What else can I do in Fiordland National Park? 44 Contact us 46 ¯ Mi lfor d P S iop ound iota hi / )" Milford k r a ¯ P Mi lfor