Hierarchies of Biocomplexity: Modeling Life’S Energetic Complexity Bradly Alicea1 1MIND Lab, Michigan State University, East Lansing, MI
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Complex Adaptive Systems Theory Applied to Virtual Scientific Collaborations: the Case of Dataone
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 8-2011 Complex adaptive systems theory applied to virtual scientific collaborations: The case of DataONE Arsev Umur Aydinoglu [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Communication Technology and New Media Commons, Library and Information Science Commons, and the Organizational Behavior and Theory Commons Recommended Citation Aydinoglu, Arsev Umur, "Complex adaptive systems theory applied to virtual scientific collaborations: The case of DataONE. " PhD diss., University of Tennessee, 2011. https://trace.tennessee.edu/utk_graddiss/1054 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Arsev Umur Aydinoglu entitled "Complex adaptive systems theory applied to virtual scientific collaborations: The case of DataONE." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Communication and Information. Suzie Allard, Major Professor We -

Biocomplexity and Conservation of Biodiversity Hotspots: Three Case Studies from the Americas J
Biocomplexity and conservation of biodiversity hotspots: three case studies from the Americas J. Baird Callicott1,*, Ricardo Rozzi1,2, Luz Delgado3, Michael Monticino4, Miguel Acevedo5 and Paul Harcombe6 1Department of Philosophy and Religion Studies, 4Department of Mathematics, and 5Department of Geography, Institute of Applied Sciences, University of North Texas, PO Box 310920, Denton, TX 76203-0920, USA 2Omora Botanical Park, Institute of Ecology and Biodiversity, University of Magallanes, Puerto Williams, Chile 3Centro de Investigaciones de Guayana, Universidad Nacional Experimental de Guayana, Puerto Ordaz, 8050 Edo. Bolı´var, Venezuela 6Department of Ecology and Evolutionary Biology, Rice University, Houston, TX 77005-1892, USA The perspective of ‘biocomplexity’ in the form of ‘coupled natural and human systems’ represents a resource for the future conservation of biodiversity hotspots in three direct ways: (i) modelling the impact on biodiversity of private land-use decisions and public land-use policies, (ii) indicating how the biocultural history of a biodiversity hotspot may be a resource for its future conservation, and (iii) identifying and deploying the nodes of both the material and psycho-spiritual connectivity between human and natural systems in service to conservation goals. Three biocomplexity case studies of areas notable for their biodiversity, selected for their variability along a latitudinal climate gradient and a human-impact gradient, are developed: the Big Thicket in southeast Texas, the Upper Botanamo River Basin in eastern Venezuela, and the Cape Horn Archipelago at the austral tip of Chile. More deeply, the biocomplexity perspective reveals alternative ways of understanding biodiversity itself, because it directs attention to the human concepts through which biodiversity is perceived and understood. -

Modeling Biocomplexity E Actors, Landscapes and Alternative Futures
Environmental Modelling & Software 22 (2006) 570e579 www.elsevier.com/locate/envsoft Modeling biocomplexity e actors, landscapes and alternative futures John P. Bolte a,*, David W. Hulse b, Stanley V. Gregory c, Court Smith d a Department of Bioengineering, Oregon State University, Oregon, USA b Department of Landscape Architecture, University of Oregon, Oregon, USA c Department of Fisheries and Wildlife, Oregon State University, Oregon, USA d Department of Anthropology, Oregon State University, Oregon, USA Received 25 October 2005; received in revised form 24 November 2005; accepted 15 December 2005 Available online 13 December 2006 Abstract Increasingly, models (and modelers) are being asked to address the interactions between human influences, ecological processes, and land- scape dynamics that impact many diverse aspects of managing complex coupled human and natural systems. These systems may be profoundly influenced by human decisions at multiple spatial and temporal scales, and the limitations of traditional process-level ecosystems modeling ap- proaches for representing the richness of factors shaping landscape dynamics in these coupled systems has resulted in the need for new analysis approaches. New tools in the areas of spatial data management and analysis, multicriteria decision-making, individual-based modeling, and com- plexity science have all begun to impact how we approach modeling these systems. The term ‘‘biocomplexity’’ has emerged as a descriptor of the rich patterns of interactions and behaviors in human and natural systems, and the challenges of analyzing biocomplex behavior is resulting in a convergence of approaches leading to new ways of understanding these systems. Important questions related to system vulnerability and resil- ience, adaptation, feedback processing, cycling, non-linearities and other complex behaviors are being addressed using models employing new representational approaches to analysis. -
Biocomplexity in the Big Thicket
Ethics, Place and Environment Vol. 9, No. 1, 21–45, March 2006 Biocomplexity in the Big Thicket J. BAIRD CALLICOTT*, MIGUEL ACEVEDO**, PETE GUNTER*, PAUL HARCOMBEy, CHRISTOPHER LINDQUIST* & MICHAEL MONTICINOz1 *Department of Philosophy, University of North Texas, TX, USA **Department of Geography, University of North Texas, TX, USA yDepartment of Ecology and Evolutionary Biology, Rice University, TX, USA zDepartment of Mathematics, University of North Texas, TX, USA Introduction: Overview of the Big Thicket and Biocomplexity The Big Thicket is an ill-defined region of southeast Texas on the coastal plain of the Gulf of Mexico between the Trinity and Sabine rivers, not far from Houston. Because the biological-diversity index of the area is one of the highest in North America, the Big Thicket National Preserve (BTNP)—an archipelago of isolated conservation ‘units’ administered by the US National Park Service—was established in 1974. The BTNP is located in a matrix of privately owned timberland, small farms, and a few small towns. The major human impacts on the region, beginning in the late 19th century and continuing into the 21st, have been logging and milling by large industrial timber operations and oil and gas extraction. Because of its proximity to the refineries of Port Arthur and the city of Beaumont and the steady increase in the availability of automobiles after World War II, residential development has also been a major impact in the region—and now represents the most potent driver of land-use/land-cover change. Few ecosystems are now free of extensive human influence. However, the way human activity affects natural systems and the way those anthropogenic changes in natural systems reciprocally affect human behavior is poorly understood. -

Biocomplexity and Conservation of Biodiversity Hotspots: Three Case Studies from the Americas J
Phil. Trans. R. Soc. B (2007) 362, 321–333 doi:10.1098/rstb.2006.1989 Published online 19 December 2006 Biocomplexity and conservation of biodiversity hotspots: three case studies from the Americas J. Baird Callicott1,*, Ricardo Rozzi1,2, Luz Delgado3, Michael Monticino4, Miguel Acevedo5 and Paul Harcombe6 1Department of Philosophy and Religion Studies, 4Department of Mathematics, and 5Department of Geography, Institute of Applied Sciences, University of North Texas, PO Box 310920, Denton, TX 76203-0920, USA 2Omora Botanical Park, Institute of Ecology and Biodiversity, University of Magallanes, Puerto Williams, Chile 3Centro de Investigaciones de Guayana, Universidad Nacional Experimental de Guayana, Puerto Ordaz, 8050 Edo. Bolı´var, Venezuela 6Department of Ecology and Evolutionary Biology, Rice University, Houston, TX 77005-1892, USA The perspective of ‘biocomplexity’ in the form of ‘coupled natural and human systems’ represents a resource for the future conservation of biodiversity hotspots in three direct ways: (i) modelling the impact on biodiversity of private land-use decisions and public land-use policies, (ii) indicating how the biocultural history of a biodiversity hotspot may be a resource for its future conservation, and (iii) identifying and deploying the nodes of both the material and psycho-spiritual connectivity between human and natural systems in service to conservation goals. Three biocomplexity case studies of areas notable for their biodiversity, selected for their variability along a latitudinal climate gradient and a human-impact gradient, are developed: the Big Thicket in southeast Texas, the Upper Botanamo River Basin in eastern Venezuela, and the Cape Horn Archipelago at the austral tip of Chile. More deeply, the biocomplexity perspective reveals alternative ways of understanding biodiversity itself, because it directs attention to the human concepts through which biodiversity is perceived and understood. -

Biocomplexity Associated with Biogeochemical Cycles in Arctic Frost-Boil Ecosystems
Biocomplexity associated with biogeochemical cycles in arctic frost-boil ecosystems Principal Investigator: Donald A. Walker Institute of Arctic Biology and Department of Biology and Wildlife, University of Alaska Fairbanks, Fairbanks, Alaska 99775, 907-474-2460, [email protected] Senior Personnel: Howard E. Epstein Department of Environmental Science, University of Virginia, Clark Hall 207, Charlottesville, VA 22904-4123, 804-924-4308, [email protected] William A. Gould International Institut of Tropical Forestry, USDA Forest Service, P.O. Box 25000, San Juan, PR 00928-02500, 787-766-5335, [email protected] William B. Krantz Department of Chemical Engineering, University of Cincinnati, Cincinnati, OH 45221-0171, 513-556-4021, [email protected] Rorik Peterson Geophysical Institute and Department of Geology and Geophysics, University of Alaska Fairbanks, Alaska, 99775, 907-474-7459, [email protected] Chien-Lu Ping Palmer Research Center, University of Alaska Fairbanks, Palmer, AK 99645, 907-746-9462, [email protected] Vladimir E. Romanovsky Geophysical Institute and Department of Geology and Geophysics, University of Alaska Fairbanks, AK, 99775, 907-474-7459, [email protected] Marilyn D. Walker Cooperative Forestry Resaerch Unit, University of Alaska Fairbanks, AK, 99775-6780, 907-474-2424, [email protected] Project Period: January 1, 2002 - December 31, 2006 Project Cost: i BIOCOMPLEXITY ASSOCIATED WITH BIOGEOCHEMICAL CYCLES IN ARCTIC FROST-BOIL ECOSYSTEMS A PROJECT SUMMARY The central goal of this project is to understand the complex linkages between biogeochemical cycles, vegetation, disturbance, and climate across the full summer temperature gradient in the Arctic in order to better predict ecosystem responses to changing climate. -

Ecological Systems As Complex Systems: Challenges for an Emerging Science
Diversity 2010, 2, 395-410; doi:10.3390/d2030395 OPEN ACCESS diversity ISSN 1424-2818 www.mdpi.com/journal/diversity Article Ecological Systems as Complex Systems: Challenges for an Emerging Science Madhur Anand 1,*, Andrew Gonzalez 2, Frédéric Guichard 2, Jurek Kolasa 3 and Lael Parrott 4 1 School of Environmental Sciences, University of Guelph, Guelph, Ontario, N1G 2W1, Canada 2 Department of Biology, McGill University, Montréal Quebec, H3A 1B1, Canada; E-Mails: [email protected] (A.G.); [email protected] (F.G.) 3 Department of Biology, McMaster University, Hamilton, Ontario, L8S 4K1, Canada; E-Mail: [email protected] 4 Département de Géographie, Université de Montréal, Montréal, Quebec, H3C 3J7, Canada; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-519-824-4210; Fax: +1-519-837-0442. Received: 30 December 2009; in revised form: 1 March 2010 / Accepted: 8 March 2010 / Published: 15 March 2010 Abstract: Complex systems science has contributed to our understanding of ecology in important areas such as food webs, patch dynamics and population fluctuations. This has been achieved through the use of simple measures that can capture the difference between order and disorder and simple models with local interactions that can generate surprising behaviour at larger scales. However, close examination reveals that commonly applied definitions of complexity fail to accommodate some key features of ecological systems, a fact that will limit the contribution of complex systems science to ecology. We highlight these features of ecological complexity—such as diversity, cross-scale interactions, memory and environmental variability—that continue to challenge classical complex systems science. -

Innovative Experiments for Sustainable Forestry Gener- Ever, There May Be Different Expectations, Timing Needs, and Ate Key Insights for the Future
BIODIVERSITY Photo by Tom Iraci Balancing Ecosystem Values Proceedings, Biodiversity Active Intentional Management (AIM) for Biodiversity and Other Forest Values Andrew B. Carey1 ABSTRACT Comparisons of natural and managed forests suggest that neither single-species management nor conventional forestry is likely to successfully meet meeting broad and diverse conservation goals. Biocomplexity is important to ecosystem function and capacity to produce useful goods and services; biocomplexity includes much more than trees of different sizes, species diversity, and individual habitat elements. Managing multiple processes of forest development, not just providing selected structures, is necessary to restore biocomplexity and ecosystem function. Experiments in inducing heterogeneity into second- growth forest canopies not only support the importance of biocomplexity to various biotic communities including soil organ- isms, vascular plants, fungi, birds, small mammals, and vertebrate predators but also suggest management can promote biocomplexity. At the landscape scale, strategies emphasizing reserves and riparian corridors that do not take into account ecological restoration of second-growth forest ecosystems and degraded streams may be self-fulfilling prophecies of forest fragmentation and landscape dysfunction. Restoring landscape function entails restoring function to second-growth forest. Intentional management can reduce the need for wide riparian buffers, produce landscapes dominated by late-seral stages that are hospitable to wildlife associated with old-growth forests, provide a sustained yield of forest products, and contribute to economic, social, and environmental sustainability. KEYWORDS: Biodiversity, northern spotted owl, northern flying squirrel, trurrles, keystone complex, mosaics. INTRODUCTION flourish concomitantly (Ray 1996). Old-growth forests are mostly in reserves and parks, distant from people, and insuf- Societies demand much of their forests. -
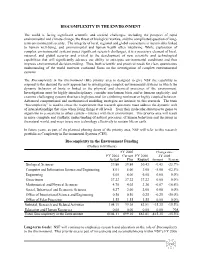
BIOCOMPLEXITY in the ENVIRONMENT Biocomplexity In
BIOCOMPLEXITY IN THE ENVIRONMENT The world is facing significant scientific and societal challenges, including the prospect of rapid environmental and climate change, the threat of biological warfare, and the complicated question of long- term environmental security. The integrity of local, regional and global ecosystems is inextricably linked to human well-being, and environmental and human health often intertwine. While exploration of complex environmental systems poses significant research challenges, it is a necessary element of local, national, and global security and critical to the development of new scientific and technological capabilities that will significantly advance our ability to anticipate environmental conditions and thus improve environmental decision-making. Thus, both scientific and practical needs for clear, quantitative understanding of the world motivate continued focus on the investigation of complex environmental systems. The Biocomplexity in the Environment (BE) priority area is designed to give NSF the capability to respond to the demand for new approaches to investigating complex environmental systems in which the dynamic behavior of biota is linked to the physical and chemical processes of the environment. Investigations must be highly interdisciplinary, consider non-human biota and/or humans explicitly, and examine challenging systems that have high potential for exhibiting nonlinear or highly coupled behavior. Advanced computational and mathematical modeling strategies are intrinsic to this research. The term “biocomplexity” is used to stress the requirement that research questions must address the dynamic web of interrelationships that arise when living things at all levels – from their molecular structures to genes to organisms to ecosystems to urban centers – interact with their environment. This priority area will result in more complete and synthetic understanding of natural processes, of human behaviors and decisions in the natural world, and ways to use new technology effectively to sustain life on earth. -

Biocomplexity in Coupled Natural–Human
Ecosystems (2005) 8: 225-232 C/*V\CVCTE?lC DOI: 10.1007/s 10021 -004-0098-7 tVvd T 0 I CM) ? 2005Springer Science+Business Media, Inc. in Biocomplexity Coupled A Natural-Human Systems: Multidimensional Framework S. T. A. Pickett,1 M. L. Cadenasso,1 and J.M. Grove2 1 Institute of Ecosystem Studies, Box AB, Millbrook, New York 12545, USA; 2Northeastern Research Station, USDA Forest Service, 705 Spear Street, P.O. Box 968, Burlington, Vermont 05401, USA Abstract As defined by Ascher, biocomplexity results from a configuration of the elements. Similarly, "multiplicity of interconnected relationships and organizational complexity increases as the focus levels/' However, no integrative framework yet shifts from unconnected units to connectivity exists to facilitate the application of this concept to among functional units. Finally, temporal com coupled human-natural systems. Indeed, the term plexity increases as the current state of a system "biocomplexity" is still used primarily as a creative comes to rely more and more on past states, and and provocative metaphor. To help advance its therefore to reflect echoes, legacies, and evolving utility, we present a framework that focuses on indirect effects of those states. This three-dimen linkages among different disciplines that are often sional, conceptual volume of biocomplexity en used in studies of coupled human-natural systems, ables connections between models that derive from including the ecological, physical, and socioeco different disciplines to be drawn at an appropriate nomic sciences. The framework consists of three level of complexity for integration. dimensions of complexity: spatial, organizational, and temporal. Spatial complexity increases as the Key words: biocomplexity; biodiversity; hetero focus changes from the type and number of the geneity; history; cross-disciplinary; integration; elements of spatial heterogeneity to an explicit space; time; organization; metaphor. -

Biocomplexity in Mangrove Ecosystems
ANRV399-MA02-15 ARI 9 November 2009 16:7 Biocomplexity in Mangrove Ecosystems I.C. Feller,1 C.E. Lovelock,2 U. Berger,3 K.L. McKee,4 S.B. Joye,5 and M.C. Ball6 1Smithsonian Environmental Research Center, Smithsonian Institution, Edgewater, Maryland 21037; email: [email protected] 2Centre for Marine Studies and School of Biological Sciences, University of Queensland, St. Lucia, QLD 4072, Australia; email: [email protected] 3Institute of Forest Growth and Computer Science, Dresden University of Technology, 01737 Tharandt, Germany; email: [email protected] 4U.S. Geological Survey-National Wetlands Research Center, Lafayette, Louisiana 70506; email: [email protected] 5Department of Marine Sciences, University of Georgia, Athens, Georgia 30602-3636; email: [email protected] 6Research School of Biological Sciences, Australian National University, Canberra ACT 0200, Australia; email: [email protected] Annu. Rev. Mar. Sci. 2010. 2:395–417 Key Words First published online as a Review in Advance on emergent properties, collective properties, trait plasticity, habitat stability, October 9, 2009 nutrient cycling, individual-based models The Annual Review of Marine Science is online at marine.annualreviews.org Abstract This article’s doi: Mangroves are an ecological assemblage of trees and shrubs adapted to grow 10.1146/annurev.marine.010908.163809 in intertidal environments along tropical coasts. Despite repeated demon- Copyright c 2010 by Annual Reviews. stration of their economic and societal value, more than 50% of the world’s All rights reserved mangroves have been destroyed, 35% in the past two decades to aquacul- 1941-1405/10/0115-0395$20.00 ture and coastal development, altered hydrology, sea-level rise, and nutri- by North Carolina State University on 01/21/14. -

Corel Ventura
A FIRST ENCOUNTER: FRENCH ENVIRONMENTAL PHILOSOPHY FROM AN ANGLO-AMERICAN PERSPECTIVE J. BAIRD CALLICOTT1 ABSTRACT. The “French exception” could be many things—language purity, cultural assimilation of immigrants, federalism counterbalanced by labor un- ionism, popular intellectualism. The French exception in environmental phi- losophy is constituted by humanism and the replacement of ethics by politics. Anglo-American environmental ethics makes of local nature a moral patient. In the French humanistic politics of global nature, global nature is indetermi- nate. Science incompletely represents global nature in both senses of the word “represents.” As an object global, nature is under-determined by a science incapable of so wide a grasp. And as subject in law, science speaks on behalf of a mute and indifferent nature, while policies regarding nature as an agent of powerful effect are decided in the political arena. KEY WORDS. French exception, ecology, environmentalism, French environ- mental philosophy, humanism, M. Serres, C. Larrère, nature, nature as politi- cal. I first encountered French environmental philosophy when I became acquainted with Catherine Larrère in 1992 at an international conference in Brazil on the eve of the Earth Summit. The conference was titled “Ethics, university, and environment.” It was organized by Fernando J. R. da Rocha, then head of the Philosophy Department at the Universidade Federal do Rio Grande do Sul in Porto Alegre. The participants were drawn from four continents—North and South America, Europe, and Australia. Larrère was one of two European participants, while the other was from Spain (Nicholás M. Sosa). As I write in July 2012, twenty years has elapsed since my first encounter with Larrère, as well as my first encounter with a distinctly French approach to environmental philoso- phy.