Week 6: Maximum Likelihood Estimation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Concentration and Consistency Results for Canonical and Curved Exponential-Family Models of Random Graphs
CONCENTRATION AND CONSISTENCY RESULTS FOR CANONICAL AND CURVED EXPONENTIAL-FAMILY MODELS OF RANDOM GRAPHS BY MICHAEL SCHWEINBERGER AND JONATHAN STEWART Rice University Statistical inference for exponential-family models of random graphs with dependent edges is challenging. We stress the importance of additional structure and show that additional structure facilitates statistical inference. A simple example of a random graph with additional structure is a random graph with neighborhoods and local dependence within neighborhoods. We develop the first concentration and consistency results for maximum likeli- hood and M-estimators of a wide range of canonical and curved exponential- family models of random graphs with local dependence. All results are non- asymptotic and applicable to random graphs with finite populations of nodes, although asymptotic consistency results can be obtained as well. In addition, we show that additional structure can facilitate subgraph-to-graph estimation, and present concentration results for subgraph-to-graph estimators. As an ap- plication, we consider popular curved exponential-family models of random graphs, with local dependence induced by transitivity and parameter vectors whose dimensions depend on the number of nodes. 1. Introduction. Models of network data have witnessed a surge of interest in statistics and related areas [e.g., 31]. Such data arise in the study of, e.g., social networks, epidemics, insurgencies, and terrorist networks. Since the work of Holland and Leinhardt in the 1970s [e.g., 21], it is known that network data exhibit a wide range of dependencies induced by transitivity and other interesting network phenomena [e.g., 39]. Transitivity is a form of triadic closure in the sense that, when a node k is connected to two distinct nodes i and j, then i and j are likely to be connected as well, which suggests that edges are dependent [e.g., 39]. -

Use of Statistical Tables
TUTORIAL | SCOPE USE OF STATISTICAL TABLES Lucy Radford, Jenny V Freeman and Stephen J Walters introduce three important statistical distributions: the standard Normal, t and Chi-squared distributions PREVIOUS TUTORIALS HAVE LOOKED at hypothesis testing1 and basic statistical tests.2–4 As part of the process of statistical hypothesis testing, a test statistic is calculated and compared to a hypothesised critical value and this is used to obtain a P- value. This P-value is then used to decide whether the study results are statistically significant or not. It will explain how statistical tables are used to link test statistics to P-values. This tutorial introduces tables for three important statistical distributions (the TABLE 1. Extract from two-tailed standard Normal, t and Chi-squared standard Normal table. Values distributions) and explains how to use tabulated are P-values corresponding them with the help of some simple to particular cut-offs and are for z examples. values calculated to two decimal places. STANDARD NORMAL DISTRIBUTION TABLE 1 The Normal distribution is widely used in statistics and has been discussed in z 0.00 0.01 0.02 0.03 0.050.04 0.05 0.06 0.07 0.08 0.09 detail previously.5 As the mean of a Normally distributed variable can take 0.00 1.0000 0.9920 0.9840 0.9761 0.9681 0.9601 0.9522 0.9442 0.9362 0.9283 any value (−∞ to ∞) and the standard 0.10 0.9203 0.9124 0.9045 0.8966 0.8887 0.8808 0.8729 0.8650 0.8572 0.8493 deviation any positive value (0 to ∞), 0.20 0.8415 0.8337 0.8259 0.8181 0.8103 0.8206 0.7949 0.7872 0.7795 0.7718 there are an infinite number of possible 0.30 0.7642 0.7566 0.7490 0.7414 0.7339 0.7263 0.7188 0.7114 0.7039 0.6965 Normal distributions. -

The Probability Lifesaver: Order Statistics and the Median Theorem
The Probability Lifesaver: Order Statistics and the Median Theorem Steven J. Miller December 30, 2015 Contents 1 Order Statistics and the Median Theorem 3 1.1 Definition of the Median 5 1.2 Order Statistics 10 1.3 Examples of Order Statistics 15 1.4 TheSampleDistributionoftheMedian 17 1.5 TechnicalboundsforproofofMedianTheorem 20 1.6 TheMedianofNormalRandomVariables 22 2 • Greetings again! In this supplemental chapter we develop the theory of order statistics in order to prove The Median Theorem. This is a beautiful result in its own, but also extremely important as a substitute for the Central Limit Theorem, and allows us to say non- trivial things when the CLT is unavailable. Chapter 1 Order Statistics and the Median Theorem The Central Limit Theorem is one of the gems of probability. It’s easy to use and its hypotheses are satisfied in a wealth of problems. Many courses build towards a proof of this beautiful and powerful result, as it truly is ‘central’ to the entire subject. Not to detract from the majesty of this wonderful result, however, what happens in those instances where it’s unavailable? For example, one of the key assumptions that must be met is that our random variables need to have finite higher moments, or at the very least a finite variance. What if we were to consider sums of Cauchy random variables? Is there anything we can say? This is not just a question of theoretical interest, of mathematicians generalizing for the sake of generalization. The following example from economics highlights why this chapter is more than just of theoretical interest. -
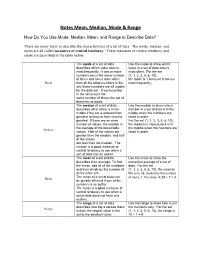
Notes Mean, Median, Mode & Range
Notes Mean, Median, Mode & Range How Do You Use Mode, Median, Mean, and Range to Describe Data? There are many ways to describe the characteristics of a set of data. The mode, median, and mean are all called measures of central tendency. These measures of central tendency and range are described in the table below. The mode of a set of data Use the mode to show which describes which value occurs value in a set of data occurs most frequently. If two or more most often. For the set numbers occur the same number {1, 1, 2, 3, 5, 6, 10}, of times and occur more often the mode is 1 because it occurs Mode than all the other numbers in the most frequently. set, those numbers are all modes for the data set. If each number in the set occurs the same number of times, the set of data has no mode. The median of a set of data Use the median to show which describes what value is in the number in a set of data is in the middle if the set is ordered from middle when the numbers are greatest to least or from least to listed in order. greatest. If there are an even For the set {1, 1, 2, 3, 5, 6, 10}, number of values, the median is the median is 3 because it is in the average of the two middle the middle when the numbers are Median values. Half of the values are listed in order. greater than the median, and half of the values are less than the median. -

Sampling Student's T Distribution – Use of the Inverse Cumulative
Sampling Student’s T distribution – use of the inverse cumulative distribution function William T. Shaw Department of Mathematics, King’s College, The Strand, London WC2R 2LS, UK With the current interest in copula methods, and fat-tailed or other non-normal distributions, it is appropriate to investigate technologies for managing marginal distributions of interest. We explore “Student’s” T distribution, survey its simulation, and present some new techniques for simulation. In particular, for a given real (not necessarily integer) value n of the number of degrees of freedom, −1 we give a pair of power series approximations for the inverse, Fn ,ofthe cumulative distribution function (CDF), Fn.Wealsogivesomesimpleandvery fast exact and iterative techniques for defining this function when n is an even −1 integer, based on the observation that for such cases the calculation of Fn amounts to the solution of a reduced-form polynomial equation of degree n − 1. We also explain the use of Cornish–Fisher expansions to define the inverse CDF as the composition of the inverse CDF for the normal case with a simple polynomial map. The methods presented are well adapted for use with copula and quasi-Monte-Carlo techniques. 1 Introduction There is much interest in many areas of financial modeling on the use of copulas to glue together marginal univariate distributions where there is no easy canonical multivariate distribution, or one wishes to have flexibility in the mechanism for combination. One of the more interesting marginal distributions is the “Student’s” T distribution. This statistical distribution was published by W. Gosset in 1908. -

Approximating the Distribution of the Product of Two Normally Distributed Random Variables
S S symmetry Article Approximating the Distribution of the Product of Two Normally Distributed Random Variables Antonio Seijas-Macías 1,2 , Amílcar Oliveira 2,3 , Teresa A. Oliveira 2,3 and Víctor Leiva 4,* 1 Departamento de Economía, Universidade da Coruña, 15071 A Coruña, Spain; [email protected] 2 CEAUL, Faculdade de Ciências, Universidade de Lisboa, 1649-014 Lisboa, Portugal; [email protected] (A.O.); [email protected] (T.A.O.) 3 Departamento de Ciências e Tecnologia, Universidade Aberta, 1269-001 Lisboa, Portugal 4 Escuela de Ingeniería Industrial, Pontificia Universidad Católica de Valparaíso, Valparaíso 2362807, Chile * Correspondence: [email protected] or [email protected] Received: 21 June 2020; Accepted: 18 July 2020; Published: 22 July 2020 Abstract: The distribution of the product of two normally distributed random variables has been an open problem from the early years in the XXth century. First approaches tried to determinate the mathematical and statistical properties of the distribution of such a product using different types of functions. Recently, an improvement in computational techniques has performed new approaches for calculating related integrals by using numerical integration. Another approach is to adopt any other distribution to approximate the probability density function of this product. The skew-normal distribution is a generalization of the normal distribution which considers skewness making it flexible. In this work, we approximate the distribution of the product of two normally distributed random variables using a type of skew-normal distribution. The influence of the parameters of the two normal distributions on the approximation is explored. When one of the normally distributed variables has an inverse coefficient of variation greater than one, our approximation performs better than when both normally distributed variables have inverse coefficients of variation less than one. -

Calculating Variance and Standard Deviation
VARIANCE AND STANDARD DEVIATION Recall that the range is the difference between the upper and lower limits of the data. While this is important, it does have one major disadvantage. It does not describe the variation among the variables. For instance, both of these sets of data have the same range, yet their values are definitely different. 90, 90, 90, 98, 90 Range = 8 1, 6, 8, 1, 9, 5 Range = 8 To better describe the variation, we will introduce two other measures of variation—variance and standard deviation (the variance is the square of the standard deviation). These measures tell us how much the actual values differ from the mean. The larger the standard deviation, the more spread out the values. The smaller the standard deviation, the less spread out the values. This measure is particularly helpful to teachers as they try to find whether their students’ scores on a certain test are closely related to the class average. To find the standard deviation of a set of values: a. Find the mean of the data b. Find the difference (deviation) between each of the scores and the mean c. Square each deviation d. Sum the squares e. Dividing by one less than the number of values, find the “mean” of this sum (the variance*) f. Find the square root of the variance (the standard deviation) *Note: In some books, the variance is found by dividing by n. In statistics it is more useful to divide by n -1. EXAMPLE Find the variance and standard deviation of the following scores on an exam: 92, 95, 85, 80, 75, 50 SOLUTION First we find the mean of the data: 92+95+85+80+75+50 477 Mean = = = 79.5 6 6 Then we find the difference between each score and the mean (deviation). -

Descriptive Statistics and Histograms Range: the Range Is a Statistic That
Descriptive Statistics and Histograms Range: The range is a statistic that helps to describe the spread of the data. The range requires numerical data to find a difference between the highest and lowest data value. Mode: The mode indicates which data value occurs most often. Median: The median describes a middle point of the data values (half the data values are above the median and half the data values are below the median). The median requires arranging the data values in ascending or descending order to determine the middle point of the data. Mean: The mean describes what would be a fair share of the data values or a balance point of the data values. How many calculators (of any kind) do you own? Range is 7; data values range from one calculator owned to 8 calculators owned. Mode is 2; the most frequent data value is 2 calculators owned. Median is 3; the middle of the data values is 3 calculators owned, or half the people surveyed own 3 or more calculators and half of the people surveyed own 3 or fewer calculators. Mean is 3.2 (to the nearest tenth); if everyone surveyed owned the same number of calculators, it would be a little more than 3 calculators, or the balance point of the data is a little more than 3 calculators owned. Possible conclusion: since the three measures of central tendency are relatively close in value, it is reasonable to conclude that the average number of calculators owned is about 3. The data value of 8 calculators is an outlier for this set of data and does pull the mean a little bit higher than the median and mode. -

3.4 Exponential Families
3.4 Exponential Families A family of pdfs or pmfs is called an exponential family if it can be expressed as ¡ Xk ¢ f(x|θ) = h(x)c(θ) exp wi(θ)ti(x) . (1) i=1 Here h(x) ≥ 0 and t1(x), . , tk(x) are real-valued functions of the observation x (they cannot depend on θ), and c(θ) ≥ 0 and w1(θ), . , wk(θ) are real-valued functions of the possibly vector-valued parameter θ (they cannot depend on x). Many common families introduced in the previous section are exponential families. They include the continuous families—normal, gamma, and beta, and the discrete families—binomial, Poisson, and negative binomial. Example 3.4.1 (Binomial exponential family) Let n be a positive integer and consider the binomial(n, p) family with 0 < p < 1. Then the pmf for this family, for x = 0, 1, . , n and 0 < p < 1, is µ ¶ n f(x|p) = px(1 − p)n−x x µ ¶ n ¡ p ¢ = (1 − p)n x x 1 − p µ ¶ n ¡ ¡ p ¢ ¢ = (1 − p)n exp log x . x 1 − p Define 8 >¡ ¢ < n x = 0, 1, . , n h(x) = x > :0 otherwise, p c(p) = (1 − p)n, 0 < p < 1, w (p) = log( ), 0 < p < 1, 1 1 − p and t1(x) = x. Then we have f(x|p) = h(x)c(p) exp{w1(p)t1(x)}. 1 Example 3.4.4 (Normal exponential family) Let f(x|µ, σ2) be the N(µ, σ2) family of pdfs, where θ = (µ, σ2), −∞ < µ < ∞, σ > 0. -

On Curved Exponential Families
U.U.D.M. Project Report 2019:10 On Curved Exponential Families Emma Angetun Examensarbete i matematik, 15 hp Handledare: Silvelyn Zwanzig Examinator: Örjan Stenflo Mars 2019 Department of Mathematics Uppsala University On Curved Exponential Families Emma Angetun March 4, 2019 Abstract This paper covers theory of inference statistical models that belongs to curved exponential families. Some of these models are the normal distribution, binomial distribution, bivariate normal distribution and the SSR model. The purpose was to evaluate the belonging properties such as sufficiency, completeness and strictly k-parametric. Where it was shown that sufficiency holds and therefore the Rao Blackwell The- orem but completeness does not so Lehmann Scheffé Theorem cannot be applied. 1 Contents 1 exponential families ...................... 3 1.1 natural parameter space ................... 6 2 curved exponential families ................. 8 2.1 Natural parameter space of curved families . 15 3 Sufficiency ............................ 15 4 Completeness .......................... 18 5 the rao-blackwell and lehmann-scheffé theorems .. 20 2 1 exponential families Statistical inference is concerned with looking for a way to use the informa- tion in observations x from the sample space X to get information about the partly unknown distribution of the random variable X. Essentially one want to find a function called statistic that describe the data without loss of im- portant information. The exponential family is in probability and statistics a class of probability measures which can be written in a certain form. The exponential family is a useful tool because if a conclusion can be made that the given statistical model or sample distribution belongs to the class then one can thereon apply the general framework that the class gives. -

A Primer on the Exponential Family of Distributions
A Primer on the Exponential Family of Distributions David R. Clark, FCAS, MAAA, and Charles A. Thayer 117 A PRIMER ON THE EXPONENTIAL FAMILY OF DISTRIBUTIONS David R. Clark and Charles A. Thayer 2004 Call Paper Program on Generalized Linear Models Abstract Generahzed Linear Model (GLM) theory represents a significant advance beyond linear regression theor,], specifically in expanding the choice of probability distributions from the Normal to the Natural Exponential Famdy. This Primer is intended for GLM users seeking a hand)' reference on the model's d]smbutional assumptions. The Exponential Faintly of D,smbutions is introduced, with an emphasis on variance structures that may be suitable for aggregate loss models m property casualty insurance. 118 A PRIMER ON THE EXPONENTIAL FAMILY OF DISTRIBUTIONS INTRODUCTION Generalized Linear Model (GLM) theory is a signtficant advance beyond linear regression theory. A major part of this advance comes from allowmg a broader famdy of distributions to be used for the error term, rather than just the Normal (Gausstan) distributton as required m hnear regression. More specifically, GLM allows the user to select a distribution from the Exponentzal Family, which gives much greater flexibility in specifying the vanance structure of the variable being forecast (the "'response variable"). For insurance apphcations, this is a big step towards more realistic modeling of loss distributions, while preserving the advantages of regresston theory such as the ability to calculate standard errors for estimated parameters. The Exponentml family also includes several d~screte distributions that are attractive candtdates for modehng clatm counts and other events, but such models will not be considered here The purpose of this Primer is to give the practicmg actuary a basic introduction to the Exponential Family of distributions, so that GLM models can be designed to best approximate the behavior of the insurance phenomenon. -

Median, Mode, and Range
5.5 Median, Mode, and Range Describe situations in real life where the mean STATES is not a good representation of the average. STANDARDS MA.6.S.6.1 1 ACTIVITY: Comparing Three Samples Work with a partner. Surveys are taken in three grade 6 –12 schools in Florida. Make up a story about the three surveys. Find the mean for each survey. Do you think the mean is a good way to represent the “average” of each group? Why or why not? a. 1112 13 14 15 16 17 18 b. 1112 13 14 15 16 17 18 c. 1112 13 14 15 16 17 18 216 Chapter 5 Ratios, Rates, and Data Analysis 2 ACTIVITY: When the Mean is Misleading Work with a partner. Read and re-read each statement. Think of a better way to represent the “average” so that the statement is not so misleading. a. Someone is trying to criticize a small high school by saying Age of Graduating Class “Last year, the average age of the 12 graduating class was 22 years old.” 10 When you look into the facts, you 8 fi nd that the class had a senior citizen who went back to school to 6 earn a diploma. Here are the ages 4 for the class. 2 18, 18, 18, 18, 18, 17, 18, 0 Number of Graduates 74 19, 18, 18, 18, 18, 18, 74 17 19 18 Age of Graduates What percent of the ages are below the mean? b. There is a small town where Families’ Yearly Income most of the people are having a diffi cult time getting by because 5 of low incomes.