Phylogeny and Vicariant Speciation of the Grey Rhebok, Pelea Capreolus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Species Fact Sheet: Roe Deer (Capreolus Capreolus) [email protected] 023 8023 7874
Species Fact Sheet: Roe Deer (Capreolus capreolus) [email protected] www.mammal.org.uk 023 8023 7874 Quick Facts Recognition: Small deer, reddish brown in summer, grey in winter. Distinctive black moustache stripe, white chin. Appears tail-less with white/ cream rump patch which is especially conspicuous when its hairs are puffed out when the deer is alarmed. Males have short antlers, erect with no more than three points. Size: Average height at shoulder 60-75cm. Males slightly larger. Weight: Adults 10—25kg Life Span: The maximum age in the wild is 16 years, but most live 7 years.. Distribution & Habitat Roe deer are widespread throughout Scotland and much of England, and in many areas they are abundant. They are increasing their range. They are spreading southwards from their Scottish refuge, and northwards and westwards from the reintroduced populations, but are not yet but are not yet established in most of the Midlands and Kent. They have never occurred in Ireland. They are generally found in open mixed, coniferous or purely deciduous woodland, particularly at edges between woodland and open habitats. Roe deer feed throughout the 24 hours, but are most active at dusk and dawn. General Ecology Behaviour Roe deer exist solitary or in small groups, with larger groups typically feeding together during the winter. At exceptionally high densities, herds of 15 or more roe deer can be seen in open fields during the spring and summer. Males are seasonally territorial, from March to August. Young females usually establish ranges close to their mothers; juvenile males are forced to disperse further afield. -

Page 5 of the 2020 Antelope, Deer and Elk Regulations
WYOMING GAME AND FISH COMMISSION Antelope, 2020 Deer and Elk Hunting Regulations Don't forget your conservation stamp Hunters and anglers must purchase a conservation stamp to hunt and fish in Wyoming. (See page 6) See page 18 for more information. wgfd.wyo.gov Wyoming Hunting Regulations | 1 CONTENTS Access on Lands Enrolled in the Department’s Walk-in Areas Elk or Hunter Management Areas .................................................... 4 Hunt area map ............................................................................. 46 Access Yes Program .......................................................................... 4 Hunting seasons .......................................................................... 47 Age Restrictions ................................................................................. 4 Characteristics ............................................................................. 47 Antelope Special archery seasons.............................................................. 57 Hunt area map ..............................................................................12 Disabled hunter season extension.............................................. 57 Hunting seasons ...........................................................................13 Elk Special Management Permit ................................................. 57 Characteristics ..............................................................................13 Youth elk hunters........................................................................ -

Boselaphus Tragocamelus</I>
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- Published Research US Geological Survey 2008 Boselaphus tragocamelus (Artiodactyla: Bovidae) David M. Leslie Jr. U.S. Geological Survey, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/usgsstaffpub Leslie, David M. Jr., "Boselaphus tragocamelus (Artiodactyla: Bovidae)" (2008). USGS Staff -- Published Research. 723. https://digitalcommons.unl.edu/usgsstaffpub/723 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- Published Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. MAMMALIAN SPECIES 813:1–16 Boselaphus tragocamelus (Artiodactyla: Bovidae) DAVID M. LESLIE,JR. United States Geological Survey, Oklahoma Cooperative Fish and Wildlife Research Unit and Department of Natural Resource Ecology and Management, Oklahoma State University, Stillwater, OK 74078-3051, USA; [email protected] Abstract: Boselaphus tragocamelus (Pallas, 1766) is a bovid commonly called the nilgai or blue bull and is Asia’s largest antelope. A sexually dimorphic ungulate of large stature and unique coloration, it is the only species in the genus Boselaphus. It is endemic to peninsular India and small parts of Pakistan and Nepal, has been extirpated from Bangladesh, and has been introduced in the United States (Texas), Mexico, South Africa, and Italy. It prefers open grassland and savannas and locally is a significant agricultural pest in India. It is not of special conservation concern and is well represented in zoos and private collections throughout the world. DOI: 10.1644/813.1. -

How Large Herbivores Subsidize Aquatic Food Webs in African
COMMENTARY Howlargeherbivoressubsidizeaquaticfoodwebsin African savannas COMMENTARY Robert M. Pringlea,1 Mass migration—the periodic, synchronized movement in the Kenyan portion of the Mara almost every year from of large numbers of animals from one place to another— 2001 to 2015, on average four to five times per year, is an important part of the life cycle of many species. resulting in a mean annual total of 6,250 wildebeest car- Such migrations are variously a means of avoiding cli- casses. These carcasses contribute more than 1,000 tons matic stress, escaping food and water scarcity, and sa- of biomass into the river—equivalent to roughly 10 blue tiating predators (thereby reducing individuals’ risk of whales—comprising dry mass of 107 tons carbon, being eaten). They are among the most spectacular 25 tons nitrogen, and 13 tons phosphorus. of natural phenomena, and also among the most Subalusky et al. (3) conducted a suite of detailed threatened: by building walls and dams, disrupting measurements and calculations to track the fate of the climate, and decimating wildlife populations, peo- these nutrients. By combining photographic surveys ple have steadily diminished and extinguished many of of carcasses with an energetic model for vultures, they the huge migrations known from historical records estimate that avian scavengers consume 4–7% of the (1, 2). Although tragic on purely aesthetic grounds— carbon and nitrogen, much of which is transported back nobody today knows the music of several million to land (Fig. 1C). Unscavenged soft tissues—such as American bison (Bison bison) snuffling and shuffling skin, muscle, and internal organs, which together make across the Great Plains—the extinction of great migra- up 56% of each carcass—decompose rapidly within tions also poses a profound threat to the functioning of 70 d, saturating the water with nutrients that are either ecosystems. -

Past Climate Changes, Population Dynamics and the Origin of Bison in Europe Diyendo Massilani1†, Silvia Guimaraes1†, Jean-Philip Brugal2,3, E
Massilani et al. BMC Biology (2016) 14:93 DOI 10.1186/s12915-016-0317-7 RESEARCHARTICLE Open Access Past climate changes, population dynamics and the origin of Bison in Europe Diyendo Massilani1†, Silvia Guimaraes1†, Jean-Philip Brugal2,3, E. Andrew Bennett1, Malgorzata Tokarska4, Rose-Marie Arbogast5, Gennady Baryshnikov6, Gennady Boeskorov7, Jean-Christophe Castel8, Sergey Davydov9, Stéphane Madelaine10, Olivier Putelat11,12, Natalia N. Spasskaya13, Hans-Peter Uerpmann14, Thierry Grange1*† and Eva-Maria Geigl1*† Abstract Background: Climatic and environmental fluctuations as well as anthropogenic pressure have led to the extinction of much of Europe’s megafauna. The European bison or wisent (Bison bonasus), one of the last wild European large mammals, narrowly escaped extinction at the onset of the 20th century owing to hunting and habitat fragmentation. Little is known, however, about its origin, evolutionary history and population dynamics during the Pleistocene. Results: Through ancient DNA analysis we show that the emblematic European bison has experienced several waves of population expansion, contraction, and extinction during the last 50,000 years in Europe, culminating in a major reduction of genetic diversity during the Holocene. Fifty-seven complete and partial ancient mitogenomes from throughout Europe, the Caucasus, and Siberia reveal that three populations of wisent (Bison bonasus)and steppe bison (B. priscus) alternately occupied Western Europe, correlating with climate-induced environmental changes. The Late Pleistocene European steppe bison originated from northern Eurasia, whereas the modern wisent population emerged from a refuge in the southern Caucasus after the last glacial maximum. A population overlap during a transition period is reflected in ca. 36,000-year-old paintings in the French Chauvet cave. -

Bantengbanteng Populationpopulation Inin Cambodia:Cambodia: Thethe Establishedestablished Baselinebaseline Densitydensity © FA / WWF-Cambodia
FACTSHEET 2011 BantengBanteng PopulationPopulation inin Cambodia:Cambodia: TheThe EstablishedEstablished BaselineBaseline DensityDensity © FA / WWF-Cambodia Between 2009-2011 in dry seasons, the research team of WWF-Cambodia conducted the first vigorous surveys on population abundance of large mammals which includes wild cattle, deer, and wild pig in the Eastern Plain Landscape (EPL) of Cambodia covering an area of approximately 6,000km2. Banteng: Globally Endangered Species Banteng (bos javanicus) is a species of wild cattle that historically inhabited deciduous and semi- evergreen forests from Northeast India and Southern Yunnan through mainland Southeast Asia and Peninsular Malaysia to Borneo and Java. Since 1996, banteng has been listed by IUCN as globally endangered on the basis of an inferred decline over the last 30 years of more than 50%. Banteng is most likely the ancestor of Southeast Asia’s domestic cattle and it is considered to be one of the most beautiful and graceful of all wild cattle species. In Cambodia, banteng populations have decreased dramatically since the late 1960s. Poaching to sell the meat and horns as trophies constitutes a major threat to remnant populations even though banteng is legally protected. © FA / WWF-Cambodia Monitoring Banteng Population in the Landscape Knowledge of animal populations is central to understanding their status and to planning their management and conservation. That is why WWF has several research projects in the EPL to gain more information about the biodiversity values of PPWS and MPF. Regular line transect surveys are conducted to collect data on large ungulates like banteng, gaur, and Eld’s deer--all potential prey species for large carnivores including tigers. -

For Vector-Borne Pathogens
Parasitology Research (2019) 118:2735–2740 https://doi.org/10.1007/s00436-019-06412-9 PROTOZOOLOGY - SHORT COMMUNICATION Screening of wild ruminants from the Kaunertal and other alpine regions of Tyrol (Austria) for vector-borne pathogens Martina Messner1 & Feodora Natalie Kayikci1 & Bita Shahi-Barogh1 & Josef Harl2 & Christian Messner3 & Hans-Peter Fuehrer1 Received: 25 June 2019 /Accepted: 25 July 2019 /Published online: 3 August 2019 # The Author(s) 2019 Abstract Knowledge about vector-borne pathogens important for human and veterinary medicine in wild ruminants in Tyrol (Austria) is scarce. Blood samples from Alpine ibex (Capra ibex; n = 44), Alpine chamois (Rupicapra rupicapra; n= 21), roe deer (Capreolus capreolus; n=18) and red deer (Cervus elaphus; n = 6) were collected over a period of 4 years (2015–2018) in four regions in North Tyrol, with a primary focus on the Kaunertal. Blood spots on filter paper were tested for the presence of DNA of vector-borne pathogens (Anaplasmataceae,Piroplasmida,Rickettsia and filarioid helminths). Anaplasma phagocytophilum and Babesia capreoli were detected in two of 89 (2.3%) blood samples. Rickettsia spp., Theileria spp. and filarioid helminths were not documented. One Alpine chamois was positive for A. phagocytophilum and B. capreoli. Moreover, an ibex from the Kaunertal region was positive for A. phagocytophilum. While the ibex was a kid less than 1 year old, the chamois was an adult individual. Further research is recommended to evaluate effects of climate change on infection rates of North Tyrolean wild ruminants by these pathogens and the distribution of their vectors. Keywords Anaplasma phagocytophilum . Austria . Babesia capreoli . Chamois . -

War and the White Rhinos
War and the White Rhinos Kai Curry-Lindahl Until 1963 the main population of the northern square-lipped (white) rhino was in the Garamba National Park, in the Congo (now Zaire) where they had increased to over 1200. That year armed rebels occupied the park, and when three years later they had been driven out, the rhinos had been drastically reduced: numbers were thought to be below 50. Dr. Curry-Iindahl describes what he found in 1966 and 1967. The northern race of the square-lipped rhinoceros Ceratotherium simum cottoni was once widely distributed in Africa north of the equator, but persecution has exterminated it over large areas. It is now known to occur only in south-western Sudan, north-eastern Congo (Kinshasa) and north-western Uganda. It is uncertain whether it still exists in northern Ubangui, in the Central African Republic. In the Sudan, where for more than ten years its range has been affected by war and serious disturbances, virtually nothing is known of its present status. In Uganda numbers dropped from about 350 in 1955 to 80 in 1962 and about 20-25 in 1969 (Cave 1963, Simon 1970); the twelve introduced into the Murchison Falls National Park in 1960, despite two being poached, increased to 18 in 1971. But the bulk of the population before 1963 was in the Garamba National Park in north-eastern Congo, in the Uele area. There, since 1938, it had been virtually undisturbed, and, thanks to the continuous research which characterised the Congo national parks before 1960, population figures are known for several periods (see the Table on page 264). -
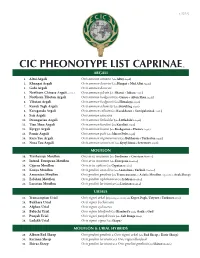
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Species of the Day: Banteng
Images © Brent Huffman / Ultimate Ungulate © Brent Huffman Species of the Day: Banteng The Banteng, Bos javanicus, is listed as ‘Endangered’ on the IUCN Red List of Threatened SpeciesTM. A species of wild cattle, the Banteng occurs in Southeast Asia from Myanmar to Indonesia, with a large introduced population in northern Australia. The Banteng has been eradicated from much of its historical range, and the remaining wild population, estimated at no more than 8,000 individuals, is continuing to decline. Habitat loss Geographical range and hunting present the greatest threats to its survival, with the illegal trade in meat and horns www.iucnredlist.org still being widespread in Southeast Asia. www.asianwildcattle.org Help Save Species Although the Banteng is legally protected across its range and occurs in a number of www.arkive.org protected areas, the natural resources of reserves in Southeast Asia often continue to be exploited. The northern Australian population may offer a conservation alternative, although genetic studies hint that the stock may originate from domesticated Bali cattle. Fortunately, a captive population is maintained worldwide which, if managed effectively and supplemented occasionally, can provide a buffer against total extinction, and offer the potential for future re- introductions into the wild. The production of the IUCN Red List of Threatened Species™ is made possible through the IUCN Red List Partnership: Species of the Day IUCN (including the Species Survival Commission), BirdLife is sponsored by International, Conservation International, NatureServe and Zoological Society of London.. -

My Somali Book, a Record of Two Shooting Trips
10 MY SOMALI BOOK to learn new things, and wonderfully bright and in- telligent. He is untiring on the march, often a reckless hunter, and will stand b}^ his master splendidly." And again, " I have made many jmigle trips in India and elsewhere, yet in no country have I had such obedient and cheerful followers and such pleasant native companions, despite their faults, as in Somali- land." This estimate is every bit as true as the other. The Somali of the interior is not, as a rule, dishonest, though he can be an exasperating liar on occasion. And he is usualty decent in his dress. Naturally indolent, he prefers to see the women do the work, but he has plenty of energy when he chooses to give it exercise, and in this respect is seen at his best on a shooting trip. And there is no doubt of his courage. A Mahomedan of the Shafai Sect, the Somali is sometimes very particular about his religious observances, but the Arab despises him as not a true Mahomedan at all. There is no occasion to detail the principal tribes, which are split up into innumerable sub-divisions. But mention must be made of the Midgdns, an outcast tribe of professional hunters. They hunt with bow and poisoned arrow, sometimes with dogs, and are wonderful trackers. As to the provision which the country makes for the hunter : the principal game to be met with in Northern Somaliland comprises Elephant, Black Rhinoceros, Lion, Leopard, Chita (hunting-leopard). MY SOMALI BOOK 11 Warthog, Ostrich, and twelve species of Antelope, to wit, Greater and Lesser Kudu, Or3^x (Beisa), Swa3aie's Hartebeest, Sommering's Gazelle (Somali-^Oi(/), Waller's Gazelle (Gerenuk), Clarke's Gazelle {Dibatag), Speke's and Pelzeln's Gazelles (both Dhero in Somali), Baira, Klipspringer {Alahlt) and Phillips' Dik-dik {Sdkdro). -

Status Report and Assessment of Wood Bison in the NWT (2016)
SPECIES STATUS REPORT Wood Bison (Bison bison athabascae) Sakāwmostos (Cree) e ta oe (Sout Slave ) e en á e ejere, t a n a n’jere ( en sųł n ) Dachan tat w ’aak’ (Teetł’ t Gw ’ n) Aak’ , a antat aak’ (Gw a Gw ’ n) Łek'a e, łuk'a e, kedä- o’, ejed (Kaska ene) Ejuda (Slavey) Tl'oo tat aak'ii, dachan tat aak'ii, akki chashuur, nin shuurchoh, nin daa ha-an (Van Tat Gw ’ n) in the Northwest Territories Threatened April 2016 Status of Wood Bison in the NWT Species at Risk Committee status reports are working documents used in assigning the status of species suspected of being at risk in the Northwest Territories (NWT). Suggested citation: Species at Risk Committee. 2016. Species Status Report for Wood Bison (Bison bison athabascae) in the Northwest Territories. Species at Risk Committee, Yellowknife, NT. © Government of the Northwest Territories on behalf of the Species at Risk Committee ISBN: 978-0-7708-0241-7 Production note: The drafts of this report were prepared by Kristi Benson (traditional and community knowledge component) and Tom Chowns (scientific knowledge component), under contract with the Government of the Northwest Territories, and edited by Claire Singer, Michelle Ramsay and Kendra McGreish. For additional copies contact: Species at Risk Secretariat c/o SC6, Department of Environment and Natural Resources P.O. Box 1320 Yellowknife, NT X1A 2L9 Tel.: (855) 783-4301 (toll free) Fax.: (867) 873-0293 E-mail: [email protected] www.nwtspeciesatrisk.ca ABOUT THE SPECIES AT RISK COMMITTEE The Species at Risk Committee was established under the Species at Risk (NWT) Act.