SMARCA5 Fusion in Extraskeletal Ewing Sarcoma&Sol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Loss of ISWI Atpase SMARCA5 (SNF2H) in Acute Myeloid Leukemia Cells Inhibits Proliferation and Chromatid Cohesion
International Journal of Molecular Sciences Article Loss of ISWI ATPase SMARCA5 (SNF2H) in Acute Myeloid Leukemia Cells Inhibits Proliferation and Chromatid Cohesion 1, 1, 1 2,3,4 1 Tomas Zikmund y , Helena Paszekova y , Juraj Kokavec , Paul Kerbs , Shefali Thakur , Tereza Turkova 1, Petra Tauchmanova 1, Philipp A. Greif 2,3,4 and Tomas Stopka 1,* 1 Biocev, 1st Medical Faculty, Charles University, 25250 Vestec, Czech Republic; [email protected] (T.Z.); [email protected] (H.P.); [email protected] (J.K.); [email protected] (S.T.); [email protected] (T.T.); [email protected] (P.T.) 2 Department of Medicine III, University Hospital, LMU Munich, D-80539 Munich, Germany; [email protected] (P.K.); [email protected] (P.A.G.) 3 German Cancer Consortium (DKTK), partner site Munich, D-80336 Munich, Germany 4 German Cancer Research Center (DKFZ), D-69120 Heidelberg, Germany * Correspondence: [email protected]; Tel.: +420-32587-3001 These authors contributed equally. y Received: 26 February 2020; Accepted: 16 March 2020; Published: 18 March 2020 Abstract: ISWI chromatin remodeling ATPase SMARCA5 (SNF2H) is a well-known factor for its role in regulation of DNA access via nucleosome sliding and assembly. SMARCA5 transcriptionally inhibits the myeloid master regulator PU.1. Upregulation of SMARCA5 was previously observed in CD34+ hematopoietic progenitors of acute myeloid leukemia (AML) patients. Since high levels of SMARCA5 are necessary for intensive cell proliferation and cell cycle progression of developing hematopoietic stem and progenitor cells in mice, we reasoned that removal of SMARCA5 enzymatic activity could affect the cycling or undifferentiated state of leukemic progenitor-like clones. -

PAX3-FOXO1 Candidate Interactors
Supplementary Table S1: PAX3-FOXO1 candidate interactors Total number of proteins: 230 Nuclear proteins : 201 Exclusive unique peptide count RH4 RMS RMS RMS Protein name Gen name FLAG#1 FLAG#2 FLAG#3 FLAG#4 Chromatin regulating complexes Chromatin modifying complexes: 6 Proteins SIN 3 complex Histone deacetylase complex subunit SAP18 SAP18 2664 CoRESt complex REST corepressor 1 RCOR1 2223 PRC1 complex E3 ubiquitin-protein ligase RING2 RNF2/RING1B 1420 MLL1/MLL complex Isoform 14P-18B of Histone-lysine N-methyltransferase MLL MLL/KMT2A 0220 WD repeat-containing protein 5 WDR5 2460 Isoform 2 of Menin MEN1 3021 Chromatin remodelling complexes: 22 Proteins CHD4/NuRD complex Isoform 2 of Chromodomain-helicase-DNA-binding protein 4 CHD4 3 21 6 0 Isoform 2 of Lysine-specific histone demethylase 1A KDM1A/LSD1a 3568 Histone deacetylase 1 HDAC1b 3322 Histone deacetylase 2 HDAC2b 96710 Histone-binding protein RBBP4 RBBP4b 10 7 6 7 Histone-binding protein RBBP7 RBBP7b 2103 Transcriptional repressor p66-alpha GATAD2A 6204 Metastasis-associated protein MTA2 MTA2 8126 SWI/SNF complex BAF SMARCA4 isoform SMARCA4/BRG1 6 13 10 0 AT-rich interactive domain-containing protein 1A ARID1A/BAF250 2610 SWI/SNF complex subunit SMARCC1 SMARCC1/BAF155c 61180 SWI/SNF complex subunit SMARCC2 SMARCC2/BAF170c 2200 Isoform 2 of SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 SMARCD1/BAF60ac 2004 Isoform 2 of SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 SMARCD3/BAF60cc 7209 -

Impaired SNF2L Chromatin Remodeling Prolongs Accessibility at Promoters Enriched for Fos/Jun Binding Sites and Delays Granule Neuron Differentiation
fnmol-14-680280 June 30, 2021 Time: 16:59 # 1 ORIGINAL RESEARCH published: 06 July 2021 doi: 10.3389/fnmol.2021.680280 Impaired SNF2L Chromatin Remodeling Prolongs Accessibility at Promoters Enriched for Fos/Jun Binding Sites and Delays Granule Neuron Differentiation Laura R. Goodwin1,2, Gerardo Zapata1,2, Sara Timpano1, Jacob Marenger1 and David J. Picketts1,2,3* 1 Regenerative Medicine Program, Ottawa Hospital Research Institute, Ottawa, ON, Canada, 2 Department of Biochemistry, Microbiology and Immunology, University of Ottawa, Ottawa, ON, Canada, 3 Cellular and Molecular Medicine, University of Ottawa, Ottawa, ON, Canada Chromatin remodeling proteins utilize the energy from ATP hydrolysis to mobilize Edited by: nucleosomes often creating accessibility for transcription factors within gene regulatory Veronica Martinez Cerdeño, University of California, Davis, elements. Aberrant chromatin remodeling has diverse effects on neuroprogenitor United States homeostasis altering progenitor competence, proliferation, survival, or cell fate. Previous Reviewed by: work has shown that inactivation of the ISWI genes, Smarca5 (encoding Snf2h) and Mitsuhiro Hashimoto, Fukushima Medical University, Japan Smarca1 (encoding Snf2l) have dramatic effects on brain development. Smarca5 Koji Shibasaki, conditional knockout mice have reduced progenitor expansion and severe forebrain Nagasaki University, Japan hypoplasia, with a similar effect on the postnatal growth of the cerebellum. In contrast, *Correspondence: Smarca1 mutants exhibited enlarged forebrains -

Downloaded Per Proteome Cohort Via the Web- Site Links of Table 1, Also Providing Information on the Deposited Spectral Datasets
www.nature.com/scientificreports OPEN Assessment of a complete and classifed platelet proteome from genome‑wide transcripts of human platelets and megakaryocytes covering platelet functions Jingnan Huang1,2*, Frauke Swieringa1,2,9, Fiorella A. Solari2,9, Isabella Provenzale1, Luigi Grassi3, Ilaria De Simone1, Constance C. F. M. J. Baaten1,4, Rachel Cavill5, Albert Sickmann2,6,7,9, Mattia Frontini3,8,9 & Johan W. M. Heemskerk1,9* Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome‑wide transcriptomes (57.8 k mRNAs). For 14.8 k protein‑coding transcripts, we assigned the proteins to 21 UniProt‑based classes, based on their preferential intracellular localization and presumed function. This classifed transcriptome‑proteome profle of platelets revealed: (i) Absence of 37.2 k genome‑ wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein‑coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43–0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identifed proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma‑derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identifed proteome of nuclear‑related, membrane and signaling proteins, as well proteins with low‑level transcripts. -

KCNMA1 Expression Is Downregulated in Colorectal Cancer Via Epigenetic Mechanisms
Article KCNMA1 Expression is Downregulated in Colorectal Cancer via Epigenetic Mechanisms Maria Sofia Basile 1, Paolo Fagone 1,*, Katia Mangano 1, Santa Mammana 2, Gaetano Magro 3, Lucia Salvatorelli 3, Giovanni Li Destri 3, Gaetano La Greca 3, Ferdinando Nicoletti 1, Stefano Puleo 3 and Antonio Pesce 3 1 Department of Biomedical and Biotechnological Sciences, University of Catania, Via S. Sofia 89, 95123 Catania, Italy; [email protected] (M.S.B.); [email protected] (K.M.); [email protected] (F.N.) 2 IRCCS Centro Neurolesi “Bonino-Pulejo”, Strada Statale 113, C.da Casazza, 98124 Messina, Italy; [email protected] 3 Department of Medical and Surgical Sciences and Advanced Technology “G.F. Ingrassia”, University of Catania, Via Santa Sofia 86, 95123 Catania, Italy; [email protected] (G.M.); [email protected] (L.S.); [email protected] (G.L.D.); [email protected] (G.L.G.); [email protected] (S.P.); [email protected] (A.P.) * Correspondence: [email protected] Received: 17 December 2018; Accepted: 16 February 2019; Published: 19 February 2019 Abstract: KCNMA1 is a gene located at 10q22 that encodes the pore-forming α-subunit of the large-conductance Ca2+-activated K+ channel. KCNMA1 is down-regulated in gastric carcinoma tumors, through hypermethylation of its promoter. In the present study, we have evaluated the expression levels of KCNMA1 both in a mouse model of Colorectal Cancer (CRC) and in human CRC samples. Additionally, epigenetic mechanisms of KCNMA1 gene regulation were investigated. We observed a significant down-regulation of KCNMA1 both in a human and mouse model of CRC. -

Identifying Proteins Bound to Native Mitotic ESC Chromosomes Reveals Chromatin Repressors Are Important for Compaction
ARTICLE https://doi.org/10.1038/s41467-020-17823-z OPEN Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction Dounia Djeghloul 1, Bhavik Patel2, Holger Kramer 3, Andrew Dimond 1, Chad Whilding4, Karen Brown1, Anne-Céline Kohler1, Amelie Feytout1, Nicolas Veland 1, James Elliott2, Tanmay A. M. Bharat 5, Abul K. Tarafder5, Jan Löwe 6, Bee L. Ng 7, Ya Guo1, Jacky Guy 8, Miles K. Huseyin 9, Robert J. Klose9, ✉ Matthias Merkenschlager 1 & Amanda G. Fisher 1 1234567890():,; Epigenetic information is transmitted from mother to daughter cells through mitosis. Here, to identify factors that might play a role in conveying epigenetic memory through cell division, we report on the isolation of unfixed, native chromosomes from metaphase-arrested cells using flow cytometry and perform LC-MS/MS to identify chromosome-bound proteins. A quantitative proteomic comparison between metaphase-arrested cell lysates and chromosome-sorted samples reveals a cohort of proteins that were significantly enriched on mitotic ESC chromosomes. These include pluripotency-associated transcription factors, repressive chromatin-modifiers such as PRC2 and DNA methyl-transferases, and proteins governing chromosome architecture. Deletion of PRC2, Dnmt1/3a/3b or Mecp2 in ESCs leads to an increase in the size of individual mitotic chromosomes, consistent with de- condensation. Similar results were obtained by the experimental cleavage of cohesin. Thus, we identify chromosome-bound factors in pluripotent stem cells during mitosis and reveal that PRC2, DNA methylation and Mecp2 are required to maintain chromosome compaction. 1 Lymphocyte Development Group, MRC London Institute of Medical Sciences, Imperial College London, Hammersmith Hospital Campus, Du Cane Road, London W12 0NN, UK. -

Blastic Plasmacytoid Dendritic Cell Neoplasm: Genomics Mark Epigenetic Dysregulation As a Primary Therapeutic Target
Myeloid Neoplasms SUPPLEMENTARY APPENDIX Blastic plasmacytoid dendritic cell neoplasm: genomics mark epigenetic dysregulation as a primary therapeutic target Maria Rosaria Sapienza, 1* Francesco Abate, 2,3* Federica Melle, 4 Stefania Orecchioni, 5 Fabio Fuligni, 6 Maryam Etebari, 1 Valentina Tabanelli, 4 Maria Antonella Laginestra, 1 Alessandro Pileri, 7,8 Giovanna Motta, 4 Maura Rossi, 1 Claudio Agostinelli, 1 Elena Sabattini, 1 Nicola Pimpinelli, 8 Mauro Truni, 9 Brunangelo Falini, 10 Lorenzo Cerroni, 11 Giovanna Ta - larico, 5 Rossana Piccioni, 12 Stefano Amente, 13 Valentina Indio, 14 Giuseppe Tarantino, 14 Francesco Brundu, 2 Marco Paulli, 15 Emilio Berti, 16 Fabio Facchetti, 17 Gaetano Ivan Dellino, 12,18 Francesco Bertolini, 5 Claudio Tripodo, 19* Raul 2,3* 4 Rabadan and Stefano A. Pileri ǂ* 1Hematopathology Unit, Department of Experimental, Diagnostic, and Specialty Medicine, S. Orsola-Malpighi Hospital, University of Bologna, Italy; 2Department of Systems Biology, Columbia University College of Physicians and Surgeons, New York, NY, USA; 3Department of Biomedical Informatics, Columbia University College of Physicians and Surgeons, New York, NY, USA; 4Division of Haematopathology, European Institute of Oncology, Milan, Italy; 5Laboratory of Hematology-Oncology, European Institute of Oncol - ogy, Milan, Italy; 6Department of Genetics and Genome Biology, The Hospital for Sick Children, Toronto, ON, Canada: 7Dermatology Unit, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Italy; 8Division of -

The Pdx1 Bound Swi/Snf Chromatin Remodeling Complex Regulates Pancreatic Progenitor Cell Proliferation and Mature Islet Β Cell
Page 1 of 125 Diabetes The Pdx1 bound Swi/Snf chromatin remodeling complex regulates pancreatic progenitor cell proliferation and mature islet β cell function Jason M. Spaeth1,2, Jin-Hua Liu1, Daniel Peters3, Min Guo1, Anna B. Osipovich1, Fardin Mohammadi3, Nilotpal Roy4, Anil Bhushan4, Mark A. Magnuson1, Matthias Hebrok4, Christopher V. E. Wright3, Roland Stein1,5 1 Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 2 Present address: Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN 3 Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN 4 Diabetes Center, Department of Medicine, UCSF, San Francisco, California 5 Corresponding author: [email protected]; (615)322-7026 1 Diabetes Publish Ahead of Print, published online June 14, 2019 Diabetes Page 2 of 125 Abstract Transcription factors positively and/or negatively impact gene expression by recruiting coregulatory factors, which interact through protein-protein binding. Here we demonstrate that mouse pancreas size and islet β cell function are controlled by the ATP-dependent Swi/Snf chromatin remodeling coregulatory complex that physically associates with Pdx1, a diabetes- linked transcription factor essential to pancreatic morphogenesis and adult islet-cell function and maintenance. Early embryonic deletion of just the Swi/Snf Brg1 ATPase subunit reduced multipotent pancreatic progenitor cell proliferation and resulted in pancreas hypoplasia. In contrast, removal of both Swi/Snf ATPase subunits, Brg1 and Brm, was necessary to compromise adult islet β cell activity, which included whole animal glucose intolerance, hyperglycemia and impaired insulin secretion. Notably, lineage-tracing analysis revealed Swi/Snf-deficient β cells lost the ability to produce the mRNAs for insulin and other key metabolic genes without effecting the expression of many essential islet-enriched transcription factors. -

Analysis and Functional Evaluation of the Hair-Cell Transcriptome
Analysis and functional evaluation of the hair-cell transcriptome Brian M. McDermott, Jr.*, Jessica M. Baucom, and A. J. Hudspeth† Howard Hughes Medical Institute and Laboratory of Sensory Neuroscience, The Rockefeller University, 1230 York Avenue, New York, NY 10021-6399 Contributed by A. J. Hudspeth, May 17, 2007 (sent for review March 31, 2007) An understanding of the molecular bases of the morphogenesis, ciated from the lagena, a receptor organ of the zebrafish’s ear. organization, and functioning of hair cells requires that the genes Linear amplification of the RNA from 200 hair cells yielded Ϸ40 expressed in these cells be identified and their functions ascer- g of aRNA, an enhancement of Ϸ1 millionfold. The resultant tained. After purifying zebrafish hair cells and detecting mRNAs labeled aRNA was hybridized to an Affymetrix microarray with oligonucleotide microarrays, we developed a subtractive (Affymetrix, Santa Clara, CA) containing Ϸ15,000 oligonucle- strategy that identified 1,037 hair cell-expressed genes whose otide probe sets. Averaging the outcomes of three experiments cognate proteins subserve functions including membrane trans- (SI Data Set 1) resulted in the identification of 6,472 transcripts port, synaptic transmission, transcriptional control, cellular adhe- scored as ‘‘present’’ (SI Data Set 2). sion and signal transduction, and cytoskeletal organization. To In the second step, we defined the transcriptome from cells of assess the validity of the subtracted hair-cell data set, we verified a nonsensory organ, the liver (SI Data Set 1). Hepatocytes were the presence of 11 transcripts in inner-ear tissue. Functional eval- selected for the subtraction process for three reasons: they are uation of two genes from the subtracted data set revealed their nonneuronal and thus unlikely to express synaptic factors; they importance in hair bundles: zebrafish larvae bearing the seahorse lack cilia (http://members.global2000.net/bowser/cilialist.html) and ift 172 mutations display specific kinociliary defects. -

Chromatin Remodeling in the UV-Induced DNA Damage Response
Chromatin remodeling in the UV-induced DNA damage response Özge Zelal Aydın Printed by: Proefschriftmaken.nl || Uitgeverij BOXPress Published by: Uitgeverij BOXPress, ’s-Hertogenbosch Cover adapted and modified by: Özge Zelal Aydın from www.health-news.com ISBN: 978-90-8891-984-8 © Copyright 2014 by Özge Zelal Aydın. All rights reserved. No part of this thesis may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, without prior written permission of the author. Chromatin remodeling in the UV-induced DNA damage response Remodellering van chromatine in de UV-geïnduceerde DNA-schade respons Proefschrift ter verkrijging van de graad van doctor aan de Erasmus Universiteit Rotterdam op gezag van de rector magnificus Prof.dr. H.A.P. Pols en volgens besluit van het College voor Promoties. De openbare verdediging zal plaatsvinden op woensdag 29 oktober 2014 om 13.30 uur door Özge Zelal Aydın geboren te Ankara, Turkije Promotiecommissie Promotor: Prof.dr. J.H.J. Hoeijmakers Overige leden: Prof.dr. A.B. Houtsmuller Dr. R.A. Poot Dr. H. van Attikum Copromotor: Prof.dr. W. Vermeulen Dr. H. Lans Aşkım da değişebilir gerçeklerim de Pırıl pırıl dalgalı bir denize karşı Yangelmişim diz boyu sulara Hepinize iyi niyetle gülümsüyorum Hiçbirinizle dövüşemem Siz ne derseniz deyiniz Benim bir gizli bildiğim var Sizin alınız al inandım Sizin morunuz mor inandım Ben tam kendime göre Ben tam dünyaya göre Ama sizin adınız ne Benim dengemi bozmayınız Turgut Uyar Contents Page Scope of the Thesis 9 Chapter I Introduction 11 Chapter -
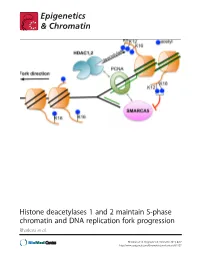
Histone Deacetylases 1 and 2 Maintain S-Phase Chromatin and DNA Replication Fork Progression Bhaskara Et Al
Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression Bhaskara et al. Bhaskara et al. Epigenetics & Chromatin 2013, 6:27 http://www.epigeneticsandchromatin.com/content/6/1/27 Bhaskara et al. Epigenetics & Chromatin 2013, 6:27 http://www.epigeneticsandchromatin.com/content/6/1/27 RESEARCH Open Access Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression Srividya Bhaskara1,2,3*, Vincent Jacques4, James R Rusche4, Eric N Olson5, Bradley R Cairns2,3 and Mahesh B Chandrasekharan1,3 Abstract Background: Histone deacetylases (HDACs) play a critical role in the maintenance of genome stability. Class I HDACs, histone deacetylase 1 and 2 (Hdac1 and Hdac2) are recruited to the replication fork by virtue of their interactions with the replication machinery. However, functions for Hdac1 and Hdac2 (Hdacs1,2) in DNA replication are not fully understood. Results: Using genetic knockdown systems and novel Hdacs1,2-selective inhibitors, we found that loss of Hdacs1,2 leads to a reduction in the replication fork velocity, and an increase in replication stress response culminating in DNA damage. These observed defects are due to a direct role for Hdacs1,2 in DNA replication, as transcription of genes involved in replication was not affected in the absence of Hdacs1,2. We found that loss of Hdacs1,2 functions increases histone acetylation (ac) on chromatin in S-phase cells and affects nascent chromatin structure, as evidenced by the altered sensitivity of newly synthesized DNA to nuclease digestion. Specifically, H4K16ac, a histone modification involved in chromatin decompaction, is increased on nascent chromatin upon abolishing Hdacs1,2 activities.