Imaging Techniques to Study Plant Virus Replication and Vertical Transmission
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Systematic, Genome-Wide Identification of Host Genes Affecting Replication of a Positive-Strand RNA Virus
Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus David B. Kushner*†, Brett D. Lindenbach*‡§, Valery Z. Grdzelishvili*, Amine O. Noueiry*, Scott M. Paul*¶, and Paul Ahlquist*‡ʈ *Institute for Molecular Virology and ‡Howard Hughes Medical Institute, University of Wisconsin, Madison, WI 53706 Contributed by Paul Ahlquist, October 23, 2003 Positive-strand RNA viruses are the largest virus class and include serves as a template for synthesis of a subgenomic (sg) mRNA, many pathogens such as hepatitis C virus and the severe acute RNA4, which encodes the viral coat protein (Fig. 1A). respiratory syndrome coronavirus (SARS). Brome mosaic virus The yeast Saccharomyces cerevisiae has proven a valuable (BMV) is a representative positive-strand RNA virus whose RNA model for normal and disease processes in human and other replication, gene expression, and encapsidation have been repro- cells. The unusual ability of BMV to direct its genomic RNA duced in the yeast Saccharomyces cerevisiae. By using traditional replication, gene expression, encapsidation, and other processes yeast genetics, host genes have been identified that function in in this yeast (7, 8) has allowed traditional yeast mutagenic controlling BMV translation, selecting BMV RNAs as replication analyses that have identified host genes involved in multiple steps templates, activating the replication complex, maintaining a lipid of BMV RNA replication and gene expression. Such host genes composition required for membrane-associated RNA replication, encode a wide variety of functions and contribute to diverse and other steps. To more globally and systematically identify such replication steps, including supporting and regulating viral trans- host factors, we used engineered BMV derivatives to assay viral lation, selecting and recruiting viral RNAs as replication RNA replication in each strain of an ordered, genome-wide set of templates, activating the RNA replication complex through yeast single-gene deletion mutants. -

Detection of Infectious Brome Mosaic Virus in Irrigation Ditches and Draining Strands in Poland
Eur J Plant Pathol https://doi.org/10.1007/s10658-018-1531-7 Detection of infectious Brome mosaic virus in irrigation ditches and draining strands in Poland Małgorzata Jeżewska & Katarzyna Trzmiel & Aleksandra Zarzyńska-Nowak Accepted: 29 June 2018 # The Author(s) 2018 Abstract Environmental waters, e.g. rivers, lakes Results confirmed the highest amino acid sequence and irrigation water, are a good source of many homology in the fragment of polymerase 2a (99.2% plant viruses. The pathogens can infect plants get- – 100%) and the most divergence in CP (96.2% - ting through damaged root hairs or small wounds 100%). This is the first report on the detection of an that appear during plant growth. First results dem- infective cereal virus in aqueous environment. onstrated common incidence of Tobacco mosaic virus (TMV) and Tomato mosaic virus (ToMV) in Keywords BMV. Water-borne virus . Cereals . RT-PCR water samples collected from irrigation ditches and drainage canals surrounding fields in Southern Greater Poland. Principal objective of this work The occurrence of plant viruses in aqueous environment was to examine if environmental water might be was studied less intensively than other water-borne vi- the source of viruses infective to cereals. The in- ruses having impact on human health. Mehle and vestigation was focused on mechanically transmit- Ravnikar (2012) thoroughly reviewed the reports and ted pathogens. Virus identification was performed listed 16 plant virus species isolated from different water by biological, electron microscopic, serological and sources, mainly from Europe, but not from Poland. molecular methods. Preliminary assays demonstrat- The main objective of our work was to fulfil this gap ed Bromemosaicvirus(BMV) infections in symp- with special attention focused on infective cereal viruses. -

The Crystallographic Structure of Brome Mosaic Virus Robertw.Lucas,Stevenb.Larsonandalexandermcpherson*
doi:10.1006/jmbi.2001.5389availableonlineathttp://www.idealibrary.comon J. Mol. Biol. (2002) 317, 95±108 The Crystallographic Structure of Brome Mosaic Virus RobertW.Lucas,StevenB.LarsonandAlexanderMcPherson* University of California, Irvine The structure of brome mosaic virus (BMV), the type member of the 560 Steinhaus Hall, Irvine bromoviridae family, has been determined from a single rhombohedral CA 92697-3900, USA crystal by X-ray diffraction, and re®ned to an R value of 0.237 for data in the range 3.4-40.0 AÊ . The structure, which represents the native, compact form at pH 5.2 in the presence of 0.1 M Mg2, was solved by molecular replacement using the model of cowpea chlorotic mottle virus (CCMV), which BMV closely resembles. The BMV model contains amino acid resi- dues 41-189 for the pentameric capsid A subunits, and residues 25-189 and 1-189 for the B and C subunits, respectively, which compose the hex- americ capsomeres. In the model there are two Mg ions and one mol- ecule of polyethylene glycol (PEG). The ®rst 25 amino acid residues of the C subunit are modeled as polyalanine. The coat protein has the cano- nical ``jellyroll'' b-barrel topology with extended amino-terminal polypep- tides as seen in other icosahedral plant viruses. Mass spectrometry shows that in native BMV virions, a signi®cant fraction of the amino-terminal peptides are apparently cleaved. No recognizable nucleic acid residue is visible in the electron density maps except at low resolution where it appears to exhibit a layered arrangement in the virion interior. -

Producing Vaccines Against Enveloped Viruses in Plants: Making the Impossible, Difficult
Review Producing Vaccines against Enveloped Viruses in Plants: Making the Impossible, Difficult Hadrien Peyret , John F. C. Steele † , Jae-Wan Jung, Eva C. Thuenemann , Yulia Meshcheriakova and George P. Lomonossoff * Department of Biochemistry and Metabolism, John Innes Centre, Norwich NR4 7UH, UK; [email protected] (H.P.); [email protected] (J.F.C.S.); [email protected] (J.-W.J.); [email protected] (E.C.T.); [email protected] (Y.M.) * Correspondence: [email protected] † Current address: Piramal Healthcare UK Ltd., Piramal Pharma Solutions, Northumberland NE61 3YA, UK. Abstract: The past 30 years have seen the growth of plant molecular farming as an approach to the production of recombinant proteins for pharmaceutical and biotechnological uses. Much of this effort has focused on producing vaccine candidates against viral diseases, including those caused by enveloped viruses. These represent a particular challenge given the difficulties associated with expressing and purifying membrane-bound proteins and achieving correct assembly. Despite this, there have been notable successes both from a biochemical and a clinical perspective, with a number of clinical trials showing great promise. This review will explore the history and current status of plant-produced vaccine candidates against enveloped viruses to date, with a particular focus on virus-like particles (VLPs), which mimic authentic virus structures but do not contain infectious genetic material. Citation: Peyret, H.; Steele, J.F.C.; Jung, J.-W.; Thuenemann, E.C.; Keywords: alphavirus; Bunyavirales; coronavirus; Flaviviridae; hepatitis B virus; human immunode- Meshcheriakova, Y.; Lomonossoff, ficiency virus; Influenza virus; newcastle disease virus; plant molecular farming; plant-produced G.P. -

RNA-Based Technologies for Engineering Plant Virus Resistance
plants Review RNA-Based Technologies for Engineering Plant Virus Resistance Michael Taliansky 1,2,*, Viktoria Samarskaya 1, Sergey K. Zavriev 1 , Igor Fesenko 1 , Natalia O. Kalinina 1,3 and Andrew J. Love 2,* 1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, 117997 Moscow, Russia; [email protected] (V.S.); [email protected] (S.K.Z.); [email protected] (I.F.); [email protected] (N.O.K.) 2 The James Hutton Institute, Invergowrie, Dundee DD2 5DA, UK 3 Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Leninskie Gory, 119991 Moscow, Russia * Correspondence: [email protected] (M.T.); [email protected] (A.J.L.) Abstract: In recent years, non-coding RNAs (ncRNAs) have gained unprecedented attention as new and crucial players in the regulation of numerous cellular processes and disease responses. In this review, we describe how diverse ncRNAs, including both small RNAs and long ncRNAs, may be used to engineer resistance against plant viruses. We discuss how double-stranded RNAs and small RNAs, such as artificial microRNAs and trans-acting small interfering RNAs, either produced in transgenic plants or delivered exogenously to non-transgenic plants, may constitute powerful RNA interference (RNAi)-based technology that can be exploited to control plant viruses. Additionally, we describe how RNA guided CRISPR-CAS gene-editing systems have been deployed to inhibit plant virus infections, and we provide a comparative analysis of RNAi approaches and CRISPR-Cas technology. The two main strategies for engineering virus resistance are also discussed, including direct targeting of viral DNA or RNA, or inactivation of plant host susceptibility genes. -

Tically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 June 2021 doi:10.20944/preprints202106.0526.v1 Review Towards the forest virome: next-generation-sequencing dras- tically expands our understanding on virosphere in temperate forest ecosystems Artemis Rumbou 1,*, Eeva J. Vainio 2 and Carmen Büttner 1 1 Faculty of Life Sciences, Albrecht Daniel Thaer-Institute of Agricultural and Horticultural Sciences, Humboldt-Universität zu Berlin, Ber- lin, Germany; [email protected], [email protected] 2 Natural Resources Institute Finland, Latokartanonkaari 9, 00790, Helsinki, Finland; [email protected] * Correspondence: [email protected] Abstract: Forest health is dependent on the variability of microorganisms interacting with the host tree/holobiont. Symbiotic mi- crobiota and pathogens engage in a permanent interplay, which influences the host. Thanks to the development of NGS technol- ogies, a vast amount of genetic information on the virosphere of temperate forests has been gained the last seven years. To estimate the qualitative/quantitative impact of NGS in forest virology, we have summarized viruses affecting major tree/shrub species and their fungal associates, including fungal plant pathogens, mutualists and saprotrophs. The contribution of NGS methods is ex- tremely significant for forest virology. Reviewed data about viral presence in holobionts, allowed us to address the role of the virome in the holobionts. Genetic variation is a crucial aspect in hologenome, significantly reinforced by horizontal gene transfer among all interacting actors. Through virus-virus interplays synergistic or antagonistic relations may evolve, which may drasti- cally affect the health of the holobiont. Novel insights of these interplays may allow practical applications for forest plant protec- tion based on endophytes and mycovirus biocontrol agents. -

The Dsrna Virus Papaya Meleira Virus and an Ssrna Virus Are Associated with Papaya Sticky Disease
RESEARCH ARTICLE The dsRNA Virus Papaya Meleira Virus and an ssRNA Virus Are Associated with Papaya Sticky Disease Tathiana Ferreira Sá Antunes1, Raquel J. Vionette Amaral1, José Aires Ventura1,2, Marcio Tadeu Godinho3, Josiane G. Amaral3, Flávia O. Souza4, Poliane Alfenas Zerbini4, Francisco Murilo Zerbini3, Patricia Machado Bueno Fernandes1* 1 Núcleo de Biotecnologia, Universidade Federal do Espírito Santo, Vitória, Espírito Santo, Brazil, 2 Instituto Capixaba de Pesquisa, Assistência Técnica e Extensão Rural, Vitória, Espírito Santo, Brazil, 3 Dep. de Fitopatologia/BIOAGRO, Universidade Federal de Viçosa, 36570–900, Viçosa, Minas Gerais, Brazil, 4 Dep. a11111 de Microbiologia/BIOAGRO, Universidade Federal de Viçosa, 36570–900, Viçosa, Minas Gerais, Brazil * [email protected] Abstract OPEN ACCESS Papaya sticky disease, or “meleira”, is one of the major diseases of papaya in Brazil and Citation: Sá Antunes TF, Amaral RJV, Ventura JA, Mexico, capable of causing complete crop loss. The causal agent of sticky disease was Godinho MT, Amaral JG, Souza FO, et al. (2016) The identified as an isometric virus with a double stranded RNA (dsRNA) genome, named dsRNA Virus Papaya Meleira Virus and an ssRNA papaya meleira virus (PMeV). In the present study, PMeV dsRNA and a second RNA band Virus Are Associated with Papaya Sticky Disease. PLoS ONE 11(5): e0155240. doi:10.1371/journal. of approximately 4.5 kb, both isolated from latex of papaya plants with severe symptoms of pone.0155240 sticky disease, were deep-sequenced. The nearly complete sequence obtained for PMeV Editor: Rogerio Margis, Universidade Federal do Rio dsRNA is 8,814 nucleotides long and contains two putative ORFs; the predicted ORF1 and Grande do Sul, BRAZIL ORF2 display similarity to capsid proteins and RdRp's, respectively, from mycoviruses ten- Totiviridae Received: January 4, 2016 tatively classified in the family . -

Synthesis and Characterization of a Full-Length Infectious Cdna Clone of Tomato Mottle Mosaic Virus
viruses Article Synthesis and Characterization of a Full-Length Infectious cDNA Clone of Tomato Mottle Mosaic Virus Liqin Tu 1,2 , Shuhua Wu 2, Danna Gao 1, Yong Liu 3, Yuelin Zhu 1,* and Yinghua Ji 2,* 1 College of Horticulture, Nanjing Agricultural University, Nanjing 210095, China; [email protected] (L.T.); [email protected] (D.G.) 2 Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences/Key Lab of Food Quality and Safety of Jiangsu Province-State Key Laboratory Breeding Base, Nanjing 210014, China; [email protected] 3 Institute of Plant Protection, Hunan Academy of Agricultural Sciences, Changsha 410125, China; [email protected] * Correspondence: [email protected] (Y.Z.); [email protected] (Y.J.); Tel.: +86-25-84396472 (Y.Z.); +86-25-84390394 (Y.J.) Abstract: Tomato mottle mosaic virus (ToMMV) is a noteworthy virus which belongs to the Virgaviridae family and causes serious economic losses in tomato. Here, we isolated and cloned the full-length genome of a ToMMV Chinese isolate (ToMMV-LN) from a naturally infected tomato (Solanum lycopersicum L.). Sequence analysis showed that ToMMV-LN contains 6399 nucleotides (nts) and is most closely related to a ToMMV Mexican isolate with a sequence identity of 99.48%. Next, an infectious cDNA clone of ToMMV was constructed by a homologous recombination approach. Both the model host N. benthamiana and the natural hosts tomato and pepper developed severe symptoms upon agroinfiltration with pToMMV, which had a strong infectivity. Electron micrographs indicated that a large number of rigid rod-shaped ToMMV virions were observed from the agroinfiltrated N. -

Sequences and Phylogenies of Plant Pararetroviruses, Viruses and Transposable Elements
Hansen and Heslop-Harrison. 2004. Adv.Bot.Res. 41: 165-193. Page 1 of 34. FROM: 231. Hansen CN, Heslop-Harrison JS. 2004 . Sequences and phylogenies of plant pararetroviruses, viruses and transposable elements. Advances in Botanical Research 41 : 165-193. Sequences and Phylogenies of 5 Plant Pararetroviruses, Viruses and Transposable Elements CELIA HANSEN AND JS HESLOP-HARRISON* DEPARTMENT OF BIOLOGY 10 UNIVERSITY OF LEICESTER LEICESTER LE1 7RH, UK *AUTHOR FOR CORRESPONDENCE E-MAIL: [email protected] 15 WEBSITE: WWW.MOLCYT.COM I. Introduction ............................................................................................................2 A. Plant genome organization................................................................................2 20 B. Retroelements in the genome ............................................................................3 C. Reverse transcriptase.........................................................................................4 D. Viruses ..............................................................................................................5 II. Retroelements........................................................................................................5 A. Viral retroelements – Retrovirales....................................................................6 25 B. Non-viral retroelements – Retrales ...................................................................7 III. Viral and non-viral elements................................................................................7 -

Crystallization of Brome Mosaic Virus and T = 1 Brome Mosaic
Virology 286, 290–303 (2001) doi:10.1006/viro.2000.0897, available online at http://www.idealibrary.com on Crystallization of Brome Mosaic Virus and T ϭ 1 Brome Mosaic Virus Particles Following a Structural Transition Robert W. Lucas, Yurii G. Kuznetsov, Steven B. Larson, and Alexander McPherson1 University of California, Irvine, Department of Molecular Biology and Biochemistry, Irvine, California 92697-3900 Received November 9, 2000; returned to author for revision January 17, 2001; accepted March 6, 2001 Brome mosaic virus (BMV), a T ϭ 3 icosahedral plant virus, can be dissociated into coat protein subunits and subunit oligomers at pH 7.5 in the presence of concentrated salts. We have found that during the course of this treatment the coat protein subunits are cleaved, presumably by plant cell proteases still present in the preparation, between amino acids 35 and 36. The truncated protein subunits will then reorganize into T ϭ 1 icosahedral particles and can be crystallized from sodium malonate. Quasi elastic light scattering and atomic force microscopy results suggest that the transition from T ϭ 3toT ϭ 1 particles can occur by separate pathways, dissociation into coat protein subunits and oligomers and reassembly into T ϭ 1 particles, or direct condensation of the T ϭ 3 virions to T ϭ 1 particles with the shedding of hexameric capsomeres. The latter process has been directly visualized using atomic force microscopy. Native T ϭ 3 virions have been crystallized in several different crystal forms, but neither a rhombohedral form nor either of two orthorhombic forms diffract beyond about 3.4 Å. -
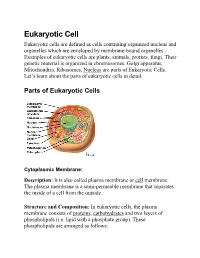
Eukaryotic Cell Eukaryotic Cells Are Defined As Cells Containing Organized Nucleus and Organelles Which Are Enveloped by Membrane-Bound Organelles
Eukaryotic Cell Eukaryotic cells are defined as cells containing organized nucleus and organelles which are enveloped by membrane-bound organelles. Examples of eukaryotic cells are plants, animals, protists, fungi. Their genetic material is organized in chromosomes. Golgi apparatus, Mitochondria, Ribosomes, Nucleus are parts of Eukaryotic Cells. Let’s learn about the parts of eukaryotic cells in detail. Parts ot Eukaryotic Cells Cytoplasmic Membrane: Description: It is also called plasma membrane or cell membrane. The plasma membrane is a semi-permeable membrane that separates the inside of a cell from the outside. Structure and Composition: In eukaryotic cells, the plasma membrane consists of proteins , carbohydrates and two layers of phospholipids (i.e. lipid with a phosphate group). These phospholipids are arranged as follows: • The polar, hydrophilic (water-loving) heads face the outside and inside of the cell. These heads interact with the aqueous environment outside and within a cell. • The non-polar, hydrophobic (water-repelling) tails are sandwiched between the heads and are protected from the aqueous environments. Scientists Singer and Nicolson(1972) described the structure of the phospholipid bilayer as the ‘Fluid Mosaic Model’. The reason is that the bi-layer looks like a mosaic and has a semi-fluid nature that allows lateral movement of proteins within the bilayer. Image: Fluid mosaic model. Orange circles – Hydrophilic heads; Lines below – Hydrophobic tails. Functions • The plasma membrane is selectively permeable i.e. it allows only selected substances to pass through. • It protects the cells from shock and injuries. • The fluid nature of the membrane allows the interaction of molecules within the membrane. -

Untersuchungen Zum Nachweis Von Pflanzenviren Mit Peptiden Und Antibody Mimics Aus Phagenbibliotheken
Untersuchungen zum Nachweis von Pflanzenviren mit Peptiden und Antibody Mimics aus Phagenbibliotheken Von der Naturwissenschaftlichen Fakultät der Gottfried Wilhelm Leibniz Universität Hannover zur Erlangung des Grades DOKTOR DER NATURWISSENSCHAFTEN Dr. rer. nat. genehmigte Dissertation von M. Sc. Dominik Lars Klinkenbuß geboren am 08.06.1986 in Dorsten 2016 Referent: Prof. Dr. Edgar Maiß Korreferent: Prof. Dr. Bernhard Huchzermeyer Tag der Promotion: 09.03.2016 KURZFASSUNG III KURZFASSUNG Trotz der kontinuierlichen Entwicklung neuerer und scheinbar fortschrittlicherer Methoden und Techniken für die Erkennung und Identifizierung von Pflanzenviren, eignen sich nur wenige dieser Methoden für Routinetests in Laboratorien. Aufgrund einzigartiger Merkmale, wie zum Beispiel die robuste Funktionalität bei einer genauen Reproduzierbarkeit, sind bis heute der enzyme-linked immunosorbent assay (ELISA) und die real-time polymerase chain reaction (qPCR) zwei der meist genutzten Diagnosetools. Das Ziel dieser Studie war die Identifikation von sogenannten „antibody mimics“ aus einer Phagenbibliothek gegen das Calibrachoa mottle virus (CbMV), Cucumber mosaic virus (CMV), Plum pox virus (PPV), Potato virus Y (PVY), Tobacco mosaic virus (TMV) und Tomato spotted wilt virus (TSWV). Im Bestfall sollen diese „antibody mimics“ die Vorteile von Antikörpern in einem ELISA besitzen und mögliche Nachteile, wie zum Beispiel die Abhängigkeit der begrenzten Ressourcen, da die benötigten Antikörper ständig nachproduziert und validiert werden müssen, vermieden werden. Dies kann durch die Produktion und Lagerung der „antibody mimics“ in Bakterienzellen erreicht werden. In einem Screeningverfahren, dem sogenannten „Biopanning“, werden Phagen selektiert, die fest an das Zielmolekül binden. In dieser Arbeit wurden diese Biopannings mit den kommerziell erhältlichen Phagenbibliotheken Ph.D.™- 12, Ph.D.™-C7C und den scFv-Bibliotheken Tomlinson I/J ausgeführt.