Lecture 3.4 Molecular Visualization Tools
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Nano-Visualization Software for Education and Research
A Nano-Visualization Software for Education and Research Lillian C. Oetting Department of Computer Science, Stanford University, Stanford, CA 94305 USA West High School, Iowa City, IA 52246 USA Tehseen Z. Raza Department of Physics and Astronomy, University of Iowa, Iowa City, IA 52242 USA Hassan Raza Department of Electrical and Computer Engineering, University of Iowa, Iowa City, IA 52242 USA Centre for Fundamental Research, Islamabad, Pakistan Abstract: We report the development of a user-friendly nano-visualization software program which can acquaint high-school students with nanotechnology. The visual introduction to atoms and molecules, which are the building blocks of this technology, is an effective way to introduce the key concepts in this area. The software’s graphical user interface enables multidimensional atomic visualization by using ball and stick schematics. Additionally, the software provides the option of wavefunction visualization for arbitrary nanomaterials and nanostructures by using extended Hückel theory. The software is instructive, application oriented and may be useful not only in high school education but also for the undergraduate research and teaching. 1 I. Introduction: The ability to accurately depict atomic, molecular and electronic structures has been a key factor in the advancement of nanotechnology. In this context, it is imperative to provide teaching and research platforms to motivate students towards this novel technology [1-3], while keeping the societal implications in perspective [4]. Nano-visualization appeals well due to its simplistic, yet elegant approach towards the visual representation of detailed concepts about quantum mechanics, quantum chemistry and linear algebra. Additionally, the conflux of quantum mechanics, numerical computation, graphical design, and computer programming gives exposure to the multi-disciplinary aspect of this technology [5-8]. -

Python Tools in Computational Chemistry (And Biology)
Python Tools in Computational Chemistry (and Biology) Andrew Dalke Dalke Scientific, AB Göteborg, Sweden EuroSciPy, 26-27 July, 2008 “Why does ‘import numpy’ take 0.4 seconds? Does it need to import 228 libraries?” - My first Numpy-discussion post (paraphrased) Your use case isn't so typical and so suffers on the import time end of the balance. - Response from Robert Kern (Others did complain. Import time down to 0.28s.) 52,000 structures PDB doubles every 2½ years HEADER PHOTORECEPTOR 23-MAY-90 1BRD 1BRD 2 COMPND BACTERIORHODOPSIN 1BRD 3 SOURCE (HALOBACTERIUM $HALOBIUM) 1BRD 4 EXPDTA ELECTRON DIFFRACTION 1BRD 5 AUTHOR R.HENDERSON,J.M.BALDWIN,T.A.CESKA,F.ZEMLIN,E.BECKMANN, 1BRD 6 AUTHOR 2 K.H.DOWNING 1BRD 7 REVDAT 3 15-JAN-93 1BRDB 1 SEQRES 1BRDB 1 REVDAT 2 15-JUL-91 1BRDA 1 REMARK 1BRDA 1 .. ATOM 54 N PRO 8 20.397 -15.569 -13.739 1.00 20.00 1BRD 136 ATOM 55 CA PRO 8 21.592 -15.444 -12.900 1.00 20.00 1BRD 137 ATOM 56 C PRO 8 21.359 -15.206 -11.424 1.00 20.00 1BRD 138 ATOM 57 O PRO 8 21.904 -15.930 -10.563 1.00 20.00 1BRD 139 ATOM 58 CB PRO 8 22.367 -14.319 -13.591 1.00 20.00 1BRD 140 ATOM 59 CG PRO 8 22.089 -14.564 -15.053 1.00 20.00 1BRD 141 ATOM 60 CD PRO 8 20.647 -15.054 -15.103 1.00 20.00 1BRD 142 ATOM 61 N GLU 9 20.562 -14.211 -11.095 1.00 20.00 1BRD 143 ATOM 62 CA GLU 9 20.192 -13.808 -9.737 1.00 20.00 1BRD 144 ATOM 63 C GLU 9 19.567 -14.935 -8.932 1.00 20.00 1BRD 145 ATOM 64 O GLU 9 19.815 -15.104 -7.724 1.00 20.00 1BRD 146 ATOM 65 CB GLU 9 19.248 -12.591 -9.820 1.00 99.00 1 1BRD 147 ATOM 66 CG GLU 9 19.902 -11.351 -10.387 1.00 99.00 1 1BRD 148 ATOM 67 CD GLU 9 19.243 -10.169 -10.980 1.00 99.00 1 1BRD 149 ATOM 68 OE1 GLU 9 18.323 -10.191 -11.782 1.00 99.00 1 1BRD 150 ATOM 69 OE2 GLU 9 19.760 -9.089 -10.597 1.00 99.00 1 1BRD 151 ATOM 70 N TRP 10 18.764 -15.737 -9.597 1.00 20.00 1BRD 152 ATOM 71 CA TRP 10 18.034 -16.884 -9.090 1.00 20.00 1BRD 153 ATOM 72 C TRP 10 18.843 -17.908 -8.318 1.00 20.00 1BRD 154 ATOM 73 O TRP 10 18.376 -18.310 -7.230 1.00 20.00 1BRD 155 . -

Chemdoodle Web Components: HTML5 Toolkit for Chemical Graphics, Interfaces, and Informatics Melanie C Burger1,2*
Burger. J Cheminform (2015) 7:35 DOI 10.1186/s13321-015-0085-3 REVIEW Open Access ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics Melanie C Burger1,2* Abstract ChemDoodle Web Components (abbreviated CWC, iChemLabs, LLC) is a light-weight (~340 KB) JavaScript/HTML5 toolkit for chemical graphics, structure editing, interfaces, and informatics based on the proprietary ChemDoodle desktop software. The library uses <canvas> and WebGL technologies and other HTML5 features to provide solutions for creating chemistry-related applications for the web on desktop and mobile platforms. CWC can serve a broad range of scientific disciplines including crystallography, materials science, organic and inorganic chemistry, biochem- istry and chemical biology. CWC is freely available for in-house use and is open source (GPL v3) for all other uses. Keywords: ChemDoodle Web Components, Chemical graphics, Animations, Cheminformatics, HTML5, Canvas, JavaScript, WebGL, Structure editor, Structure query Introduction Mobile browsers did support HTML5, which opened How we communicate chemical information is increas- the door to web applications built with only HTML, ingly technology driven. Learning management systems, CSS and JavaScript (JS), such as the ChemDoodle Web virtual classrooms and MOOCs are a few examples where Components. chemistry educators need forward compatible tools for digital natives. Companies that implement emerg- Review ing web technologies can find efficiencies and benefit The ChemDoodle Web Components technology stack from competitive advantages. The first chemical graph- and features ics toolkit for the web, MDL Chime, was introduced in The ChemDoodle Web Components library, released in 1996 [1]. Based on the molecular visualization program 2009, is the first chemistry toolkit for structure viewing RasMol, Chime was developed as a plugin for Netscape and editing that is originally built using only web stand- and later for Internet Explorer and Firefox. -
Instructions for PDB Downloading (From Either Website)
Molecular Visualization A BBSI Tutorial http://www.ccbb.pitt.edu/BBSI/index.htm By: Jeffry D. Madura Joshua A. Plumley Thomas J. Dick Table of Contents Instructions for PDB downloading..........................................................2 RasMol Tutorial ...........................................................................................3 VMD Tutorial ...............................................................................................5 CAChe Tutuorial..........................................................................................8 MOE Tutorial ............................................................................................. 10 Chimera Tutorial ....................................................................................... 13 Exercises .................................................................................................... 16 1 Instructions for PDB downloading (from either website) -Go to: - Go to: www.rcsb.org/pdb/ www.pdb.bu.edu/oca-bin/pdblite -Type in name of protein (examples at - Type in name of Protein / bottom of the page). Macromolecule ( examples at bottom of page). - Click on Name icon (first name in purple box). - Click on Retrieve Released Data Matching Your Query icon. -On the left side of the screen, click on Download/Display Structure - Highlight any of the molecule name and click on the View/ Analyze/ -Under Download the Structure File, Save Macro Molecule icon. right click on the X where the PDB(top) meets with none, under - Under the Data Retrieval section, -

USER MANUAL Version 4.5
GROMACS Groningen Machine for Chemical Simulations USER MANUAL Version 4.5 GROMACS USER MANUAL Version 4.5 Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen, Aldert van Buuren, Par¨ Bjelkmar, Rudi van Drunen, Anton Feenstra, Gerrit Groenhof, Peter Kasson, Per Larsson, Peiter Meulenhoff, Teemu Murtola, Szilard´ Pall,´ Sander Pronk, Roland Schultz, Michael Shirts, Alfons Sijbers, Peter Tieleman Berk Hess, David van der Spoel, and Erik Lindahl. Additional contributions by Mark Abraham, Christoph Junghans, Carsten Kutzner, Justin A. Lemkul, Erik Marklund, Maarten Wolf. c 1991–2000: Department of Biophysical Chemistry, University of Groningen. Nijenborgh 4, 9747 AG Groningen, The Netherlands. c 2001–2010: The GROMACS development teams at the Royal Institute of Technology and Uppsala University, Sweden. More information can be found on our website: www.gromacs.org. iv Preface & Disclaimer This manual is not complete and has no pretention to be so due to lack of time of the contributors – our first priority is to improve the software. It is worked on continuously, which in some cases might mean the information is not entirely correct. Comments are welcome, please send them by e-mail to [email protected], or to one of the mailing lists (see www.gromacs.org). We try to release an updated version of the manual whenever we release a new version of the soft- ware, so in general it is a good idea to use a manual with the same major and minor release number as your GROMACS installation. Any revision numbers (like 3.1.1) are however independent, to make it possible to implement bug fixes and manual improvements if necessary. -
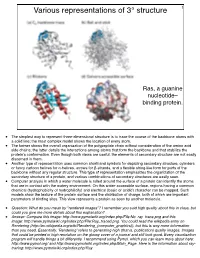
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

Optimizing the Use of Open-Source Software Applications in Drug
DDT • Volume 11, Number 3/4 • February 2006 REVIEWS TICS INFORMA Optimizing the use of open-source • software applications in drug discovery Reviews Werner J. Geldenhuys1, Kevin E. Gaasch2, Mark Watson2, David D. Allen1 and Cornelis J.Van der Schyf1,3 1Department of Pharmaceutical Sciences, School of Pharmacy,Texas Tech University Health Sciences Center, Amarillo,TX, USA 2West Texas A&M University, Canyon,TX, USA 3Pharmaceutical Chemistry, School of Pharmacy, North-West University, Potchefstroom, South Africa Drug discovery is a time consuming and costly process. Recently, a trend towards the use of in silico computational chemistry and molecular modeling for computer-aided drug design has gained significant momentum. This review investigates the application of free and/or open-source software in the drug discovery process. Among the reviewed software programs are applications programmed in JAVA, Perl and Python, as well as resources including software libraries. These programs might be useful for cheminformatics approaches to drug discovery, including QSAR studies, energy minimization and docking studies in drug design endeavors. Furthermore, this review explores options for integrating available computer modeling open-source software applications in drug discovery programs. To bring a new drug to the market is very costly, with the current of combinatorial approaches and HTS. The addition of computer- price tag approximating US$800 million, according to data reported aided drug design technologies to the R&D approaches of a com- in a recent study [1]. Therefore, it is not surprising that pharma- pany, could lead to a reduction in the cost of drug design and ceutical companies are seeking ways to optimize costs associated development by up to 50% [6,7]. -

VMD User's Guide
VMD User’s Guide Version 1.9.3 November 27, 2016 NIH Biomedical Research Center for Macromolecular Modeling and Bioinformatics Theoretical and Computational Biophysics Group1 Beckman Institute for Advanced Science and Technology University of Illinois at Urbana-Champaign 405 N. Mathews Urbana, IL 61801 http://www.ks.uiuc.edu/Research/vmd/ Description The VMD User’s Guide describes how to run and use the molecular visualization and analysis program VMD. This guide documents the user interfaces displaying and grapically manipulating molecules, and describes how to use the scripting interfaces for analysis and to customize the be- havior of VMD. VMD development is supported by the National Institutes of Health grant numbers NIH 9P41GM104601 and 5R01GM098243-02. 1http://www.ks.uiuc.edu/ Contents 1 Introduction 10 1.1 Contactingtheauthors. ....... 11 1.2 RegisteringVMD.................................. 11 1.3 CitationReference ............................... ...... 11 1.4 Acknowledgments................................. ..... 12 1.5 Copyright and Disclaimer Notices . .......... 12 1.6 For information on our other software . .......... 14 2 Hardware and Software Requirements 16 2.1 Basic Hardware and Software Requirements . ........... 16 2.2 Multi-core CPUs and GPU Acceleration . ......... 16 2.3 Parallel Computing on Clusters and Supercomputers . .............. 17 3 Tutorials 18 3.1 RapidIntroductiontoVMD. ...... 18 3.2 Viewing a molecule: Myoglobin . ........ 18 3.3 RenderinganImage ................................ 20 3.4 AQuickAnimation................................. 20 3.5 An Introduction to Atom Selection . ......... 21 3.6 ComparingTwoStructures . ...... 21 3.7 SomeNiceRepresenations . ....... 22 3.8 Savingyourwork.................................. 23 3.9 Tracking Script Command Versions of the GUI Actions . ............ 23 4 Loading A Molecule 25 4.1 Notes on common molecular file formats . ......... 25 4.2 Whathappenswhenafileisloaded? . ....... 26 4.3 Babelinterface ................................. -

Computational Chemistry: a Practical Guide for Applying Techniques to Real-World Problems
Computational Chemistry: A Practical Guide for Applying Techniques to Real-World Problems. David C. Young Copyright ( 2001 John Wiley & Sons, Inc. ISBNs: 0-471-33368-9 (Hardback); 0-471-22065-5 (Electronic) COMPUTATIONAL CHEMISTRY COMPUTATIONAL CHEMISTRY A Practical Guide for Applying Techniques to Real-World Problems David C. Young Cytoclonal Pharmaceutics Inc. A JOHN WILEY & SONS, INC., PUBLICATION New York . Chichester . Weinheim . Brisbane . Singapore . Toronto Designations used by companies to distinguish their products are often claimed as trademarks. In all instances where John Wiley & Sons, Inc., is aware of a claim, the product names appear in initial capital or all capital letters. Readers, however, should contact the appropriate companies for more complete information regarding trademarks and registration. Copyright ( 2001 by John Wiley & Sons, Inc. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic or mechanical, including uploading, downloading, printing, decompiling, recording or otherwise, except as permitted under Sections 107 or 108 of the 1976 United States Copyright Act, without the prior written permission of the Publisher. Requests to the Publisher for permission should be addressed to the Permissions Department, John Wiley & Sons, Inc., 605 Third Avenue, New York, NY 10158-0012, (212) 850-6011, fax (212) 850-6008, E-Mail: PERMREQ @ WILEY.COM. This publication is designed to provide accurate and authoritative information in regard to the subject matter covered. It is sold with the understanding that the publisher is not engaged in rendering professional services. If professional advice or other expert assistance is required, the services of a competent professional person should be sought. -

Open Source Molecular Modeling
Accepted Manuscript Title: Open Source Molecular Modeling Author: Somayeh Pirhadi Jocelyn Sunseri David Ryan Koes PII: S1093-3263(16)30118-8 DOI: http://dx.doi.org/doi:10.1016/j.jmgm.2016.07.008 Reference: JMG 6730 To appear in: Journal of Molecular Graphics and Modelling Received date: 4-5-2016 Accepted date: 25-7-2016 Please cite this article as: Somayeh Pirhadi, Jocelyn Sunseri, David Ryan Koes, Open Source Molecular Modeling, <![CDATA[Journal of Molecular Graphics and Modelling]]> (2016), http://dx.doi.org/10.1016/j.jmgm.2016.07.008 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. Open Source Molecular Modeling Somayeh Pirhadia, Jocelyn Sunseria, David Ryan Koesa,∗ aDepartment of Computational and Systems Biology, University of Pittsburgh Abstract The success of molecular modeling and computational chemistry efforts are, by definition, de- pendent on quality software applications. Open source software development provides many advantages to users of modeling applications, not the least of which is that the software is free and completely extendable. In this review we categorize, enumerate, and describe available open source software packages for molecular modeling and computational chemistry. 1. Introduction What is Open Source? Free and open source software (FOSS) is software that is both considered \free software," as defined by the Free Software Foundation (http://fsf.org) and \open source," as defined by the Open Source Initiative (http://opensource.org). -

Download PDF 137.14 KB
Why should biochemistry students be introduced to molecular dynamics simulations—and how can we introduce them? Donald E. Elmore Department of Chemistry and Biochemistry Program; Wellesley College; Wellesley, MA 02481 Abstract Molecular dynamics (MD) simulations play an increasingly important role in many aspects of biochemical research but are often not part of the biochemistry curricula at the undergraduate level. This article discusses the pedagogical value of exposing students to MD simulations and provides information to help instructors consider what software and hardware resources are necessary to successfully introduce these simulations into their courses. In addition, a brief review of the MD based activities in this issue and other sources are provided. Keywords molecular dynamics simulations, molecular modeling, biochemistry education, undergraduate education, computational chemistry Published in Biochemistry and Molecular Biology Education, 2016, 44:118-123. Several years ago, I developed a course on the modeling of biochemical systems at Wellesley College, which was described in more detail in a previous issue of this journal [1]. As I set out to design that course, I decided to place a particular emphasis on molecular dynamics (MD) simulations. In full candor, this decision was likely related to my experience integrating these simulations into my own research, but I also believe it was due to some inherent advantages of exposing students to this particular computational approach. Notably, even as this initial biochemical molecular modeling course evolved into a more general (i.e. not solely biochemical) course in computational chemistry, a significant emphasis on MD has remained, emphasizing the central role I and my other colleague who teaches the current course felt it played in students’ education. -

Ballview a Molecular Viewer and Modeling Tool
BALLView A molecular viewer and modeling tool Dissertation zur Erlangung des Grades des Doktors der Ingenieurwissenschaften der Naturwissenschaftlich– Technischen Fakultäten der Universität des Saarlandes vorgelegt von Diplom-Biologe Andreas Moll Saarbrücken im Mai 2007 Tag des Kolloquiums: 18. Juli 2007 Dekan: Prof. Dr. Thorsten Herfet Mitglieder des Prüfungsausschusses: Prof. Dr. Philipp Slusallek Prof. Dr. Hans-Peter Lenhof Prof. Dr. Oliver Kohlbacher Dr. Dirk Neumann Acknowledgments The work on this thesis was carried out during the years 2002-2007 at the Center for Bioinformatics in the group of Prof. Dr. Hans-Peter Lenhof who also was the supervisor of the thesis. With his deeply interesting lecture on bioinformatics, Prof. Dr. Hans-Peter Lenhof kindled my interest in this field and gave me the freedom to do research in those areas that fascinated me most. The implementation of BALLView would have been unthinkable without the help of all the people who contributed code and ideas. In particular, I want to thank Prof. Dr. Oliver Kohlbacher for his splendid work on the BALL library, on which this thesis is based on. Furthermore, Prof. Kohlbacher had at any time an open ear for my questions. Next I want to thank Dr. Andreas Hildebrandt, who had good advices for the majority of problems that I was confronted with. In addition he contributed code for database access, field line calculations, spline points calculations, 2D depiction of molecules, and for the docking in- terface. Heiko Klein wrote the precursor of the VIEW library. Anne Dehof implemented the peptide builder, the secondary structure assignment, and a first version of the editing mo- de.