How Tarzan Works - Some Information on The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Federal Zone: Cracking the Code of Internal Revenue Eleventh Edition by Paul Andrew Mitchell, B.A., M.S. Counselor at Law, F
The Federal Zone: Cracking the Code of Internal Revenue Eleventh Edition by Paul Andrew Mitchell, B.A., M.S. Counselor at Law, Federal Witness, and Private Attorney General Published by Supreme Law Publishers c/o Forwarding Agent 501 W. Broadway, Suite A-332 San Diego 92101 CALIFORNIA, USA March 1, 2001 A.D. The Federal Zone: Disclaimer This book is designed to educate you about federal income tax law, the Treasury regulations which promulgate that law, and the various court decisions which have interpreted both. It is sold with the understanding that the Author and Publisher are not engaged in rendering legal services of any kind. The right to author and publish this book, no matter how often the statutes, regulations and case law are quoted, is explicitly guaranteed by the First Amendment to the Constitution for the United States of America, a written contract to which the federal government, the 50 States, and their respective agencies are all parties. Federal and State laws are changing constantly, and no single book can possibly address all legal situations in which you may find yourself, now or in the future. The Federal Zone: Cracking the Code of Internal Revenue Common Law Copyright March 1, 2001 A.D. Paul Andrew Mitchell, B.A., M.S. Counselor at Law, Federal Witness, and Private Attorney General The information contained in this book is lawfully protected from copyright violations and reproduction infringements of any kind. Violators are hereby warned that they can and will be prosecuted to the full extent of American law, at the sole discretion of the Author and Publisher. -

Tarzan in the Early-20Th Century French Fantasy Landscape By
Wesleyan University The Honors College The Missing Link: Tarzan in the Early-20th Century French Fantasy Landscape by Medha Swaminathan Class of 2019 A thesis submitted to the faculty of Wesleyan University in partial fulfillment of the requirements for the Degree of Bachelor of Arts with Departmental Honors in French Studies Middletown, Connecticut April, 2019 Table of Contents Introduction ................................................................................................................... 1 Embracing the Invented in the “Benevolent” Colonial ................................................ 9 Imagining “Africa” ..................................................................................................... 19 Le Tour du Monde en Un Jour: Tarzan and the 1930s Paris Colonial Exhibitions .... 36 “Civilization” vs. “Civilized” vs. “Savage” ................................................................ 49 Homme Idéal or Missing Link? Fetish, Fascination, and Fear in French Eugenics ... 57 Sex, Youth, Beauty, Valor, and the Légionnaire ........................................................ 70 Saturnin Farandoul: Tarzan’s French Foil? ................................................................ 81 “Comment dit-on sites de rêve en anglais ?” .............................................................. 96 References ................................................................................................................. 100 Acknowledgements This project would not have been possible without an incredible amount -

The Panama Canal Review Is Published Twice a Year
UNIVERSITY OF FLORIDA LIBRARIES m.• #.«, I i PANAMA w^ p IE I -.a. '. ±*L. (Qfx m Uu *£*£ - Willie K Friar David S. Parker Editor, English Edition Governor-President Jose T. Tunon Charles I. McGinnis Editor, Spanish Edition Lieutenant Governor Writers Eunice Richard, Frank A. Baldwin Fannie P. Hernandez, Publication Franklin Castrellon and Dolores E. Suisman Panama Canal Information Officer Official Panama Canal the Review will be appreciated. Review articles may be reprinted without further clearance. Credit tu regular mail airmail $2, single copies 50 cents. The Panama Canal Review is published twice a year. Yearly subscription: $1, Canal Company, to Panama Canal Review, Box M, Balboa Heights, C.Z. For subscription, send check or money order, made payable to the Panama Editorial Office is located in Room 100, Administration Building, Balboa Heights, C.Z. Printed at the Panama Canal Printing Plant, La Boca, C.Z. Contents Our Cover The Golden Huacas of Panama 3 Huaca fanciers will find their favor- the symbolic characters of Treasures of a forgotten ites among the warrior, rainbow, condor god, eagle people arouse the curiosity and alligator in this display of Pan- archeologists around the of ama's famous golden artifacts. world. The huacas, copied from those recov- Snoopy Speaks Spanish 8 ered from the graves of pre-Columbian loaned to The In the phonetics of the fun- Carib Indians, were Review by Neville Harte. The well nies, a Spanish-speaking dog known local archeologist also provided doesn't say "bow wow." much of the information for the article Balseria 11 from his unrivaled knowledge of the Broken legs are the name of subject—the fruit of a 26-year-long love affair with the huaca, and the country the game when the Guaymis and people of Panama, past and present. -

Zone – Acte III – Questions De Compréhension
Zone – Acte III – Questions de compréhension 1. Quand Ciboulette demande à Moineau si son livre finit bien, il dit non. Expliquez la fin de son histoire. Pourquoi pensez-vous qu’il n’aime pas la fin? - Moineau explique que « Les méchants » se font punir par les « bons ». Il ne l’aime pas parce que, dans les yeux de la société, lui et son groupe sont « les méchants » de même qu’ils avaient tous leurs propres raisons de vendre les cigarettes, et autre que Tarzan, ils n’ont fait rien de mal à personne d’autre. 2. Passe-Partout utilise quel moyen pour aider à convaincre Moineau et Ciboulette de lui suivre comme nouveau chef? - Il indique que Tit-Noir avait bien caché leur argent et que la police ne l’a pas trouvé, donc il a pris le tout et il explique qu’il va seulement redonner l’argent aux autres s’ils lui acceptent comme nouveau chef. Ciboulette et Moineau ne sont pas en accord. 3. A) Pourquoi Passe-Partout sort à l’entrée de Tit-Noir? - Tit-Noir entre et annonce que Tarzan s’est évadé des policiers, le groupe assume qu’il reviendra bientôt au hangar. Passe-Partout, sachant qu’il a trahi leur chef, quitte avant que Tarzan arrive afin d’éviter une réunion malheureux avec lui. B) Quel est le problème avec la disparition de Passe-Partout? - Pour que le reste du groupe puisse aider Tarzan à se cacher de la police, il faut avoir de l’argent pour vivre. Passe-Partout vient de quitter avec tout leur argent. -

David Hare, Surrealism, and the Comics Mona Hadler Brooklyn College and the Graduate Center, CUNY
93 David Hare, Surrealism, and the Comics Mona Hadler Brooklyn College and The Graduate Center, CUNY The history of the comic book in the United States has been closely allied with mass culture debates ranging from Clement Greenberg’s 1939 essay “Avant-Garde and Kitsch” to Fredric Wertham’s attack on the comic book industry in his now infamous 1954 book, Seduction of the Innocent. An alternative history can be constructed, however, by linking the comics to the expatriate Surrealist community in New York in the thirties and early forties with its focus on fantasy, dark humor, the poetics and ethics of evil, transgressive and carnivalesque body images, and restless desire. Surrealist artists are well known for their fascination with black humor, pulp fi ction, and crime novels—in particular the popular series Fantômas. Indeed, the image of the scoundrel in elegant attire holding a bloody dagger, featured in the writings of the Surrealist Robert Desnos (Color Plate 10), epitomized this darkling humor (Desnos 377-79).1 Pictures of Fantômas and articles on the pulp market peppered the pages of Surrealist magazines in Europe and America, from Documents in France (1929-1930) to VVV and View in New York in the early forties. By moving the discussion of the comics away from a Greenbergian or Frankfurt school discourse on mass culture to the debates surrounding Surrealism, other issues come to the fore. The comics become a site for the exploration of mystery, the imagination, criminality, and freedom. The comic book industry originated in the United States in the late thirties and early forties on the heels of a booming pulp fi ction market. -

Comic Book Collection
2008 preview: fre comic book day 1 3x3 Eyes:Curse of the Gesu 1 76 1 76 4 76 2 76 3 Action Comics 694/40 Action Comics 687 Action Comics 4 Action Comics 7 Advent Rising: Rock the Planet 1 Aftertime: Warrior Nun Dei 1 Agents of Atlas 3 All-New X-Men 2 All-Star Superman 1 amaze ink peepshow 1 Ame-Comi Girls 4 Ame-Comi Girls 2 Ame-Comi Girls 3 Ame-Comi Girls 6 Ame-Comi Girls 8 Ame-Comi Girls 4 Amethyst: Princess of Gemworld 9 Angel and the Ape 1 Angel and the Ape 2 Ant 9 Arak, Son of Thunder 27 Arak, Son of Thunder 33 Arak, Son of Thunder 26 Arana 4 Arana: The Heart of the Spider 1 Arana: The Heart of the Spider 5 Archer & Armstrong 20 Archer & Armstrong 15 Aria 1 Aria 3 Aria 2 Arrow Anthology 1 Arrowsmith 4 Arrowsmith 3 Ascension 11 Ashen Victor 3 Astonish Comics (FCBD) Asylum 6 Asylum 5 Asylum 3 Asylum 11 Asylum 1 Athena Inc. The Beginning 1 Atlas 1 Atomic Toybox 1 Atomika 1 Atomika 3 Atomika 4 Atomika 2 Avengers Academy: Fear Itself 18 Avengers: Unplugged 6 Avengers: Unplugged 4 Azrael 4 Azrael 2 Azrael 2 Badrock and Company 3 Badrock and Company 4 Badrock and Company 5 Bastard Samurai 1 Batman: Shadow of the Bat 27 Batman: Shadow of the Bat 28 Batman:Shadow of the Bat 30 Big Bruisers 1 Bionicle 22 Bionicle 20 Black Terror 2 Blade of the Immortal 3 Blade of the Immortal unknown Bleeding Cool (FCBD) Bloodfire 9 bloodfire 9 Bloodshot 2 Bloodshot 4 Bloodshot 31 bloodshot 9 bloodshot 4 bloodshot 6 bloodshot 15 Brath 13 Brath 12 Brath 14 Brigade 13 Captain Marvel: Time Flies 4 Caravan Kidd 2 Caravan Kidd 1 Cat Claw 1 catfight 1 Children of -
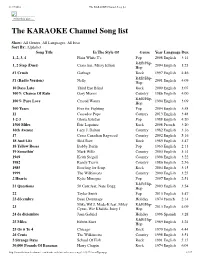
The KARAOKE Channel Song List
11/17/2016 The KARAOKE Channel Song list Print this List ... The KARAOKE Channel Song list Show: All Genres, All Languages, All Eras Sort By: Alphabet Song Title In The Style Of Genre Year Language Dur. 1, 2, 3, 4 Plain White T's Pop 2008 English 3:14 R&B/Hip- 1, 2 Step (Duet) Ciara feat. Missy Elliott 2004 English 3:23 Hop #1 Crush Garbage Rock 1997 English 4:46 R&B/Hip- #1 (Radio Version) Nelly 2001 English 4:09 Hop 10 Days Late Third Eye Blind Rock 2000 English 3:07 100% Chance Of Rain Gary Morris Country 1986 English 4:00 R&B/Hip- 100% Pure Love Crystal Waters 1994 English 3:09 Hop 100 Years Five for Fighting Pop 2004 English 3:58 11 Cassadee Pope Country 2013 English 3:48 1-2-3 Gloria Estefan Pop 1988 English 4:20 1500 Miles Éric Lapointe Rock 2008 French 3:20 16th Avenue Lacy J. Dalton Country 1982 English 3:16 17 Cross Canadian Ragweed Country 2002 English 5:16 18 And Life Skid Row Rock 1989 English 3:47 18 Yellow Roses Bobby Darin Pop 1963 English 2:13 19 Somethin' Mark Wills Country 2003 English 3:14 1969 Keith Stegall Country 1996 English 3:22 1982 Randy Travis Country 1986 English 2:56 1985 Bowling for Soup Rock 2004 English 3:15 1999 The Wilkinsons Country 2000 English 3:25 2 Hearts Kylie Minogue Pop 2007 English 2:51 R&B/Hip- 21 Questions 50 Cent feat. Nate Dogg 2003 English 3:54 Hop 22 Taylor Swift Pop 2013 English 3:47 23 décembre Beau Dommage Holiday 1974 French 2:14 Mike WiLL Made-It feat. -

Fantasy Videos 4 Star #670DCC27
267 2001: A Space Odyssey (25th anniversary) 4054 8 1/2 (Italian/Subtitled B&W) 9219 After Life (Japanese/Subtitled) 5965 Animation...Winsor McCay (Silent cartoons inc./Gertie) 7080 Babe (Pig) (1995) 8609 Babe: Pig in the City 4985 Beauty and the Beast (1946 French/Subtitled B&W) 5031 Beauty and the Beast (Disney animated 1991) 5966 Cameraman’s Revenge & Other Fantastic Tales (Silent) 626 Christmas Carol (1951 B&W) (aka Scrooge) 9559 Crouching Tiger, Hidden Dragon (Chinese/Subtitled) 7946 Empire Strikes Back (1997 Special Edition) 153 Empire Strikes Back (remastered edition) 9405 Fantasticks (1995) 3041 Field of Dreams 6874 Films of Kenneth Anger: Eaux d’Artifice...(1953) 10705 Finding Nemo 9332 Golden Age (L’Age D’Or) (B&W French/Subtitled) 10748 Halloween (widescreen extended version) 10128 Harry Potter and the Sorcerer’s Stone 3202 Harvey (B&W) 3044 Heaven Can Wait (1943) 3927 Juliet of the Spirits (Italian/Subtitled) 6516 Leolo (French/Subtitled) 1937 Lost Horizon (1937 B&W) 10654 Lord of the Rings: The Two Towers (2002) 230 E. North St. Sidney, Ohio 45365 10713 Lord of the Rings: The Two Towers (2002) (extended version) (937) 492-8354 1174 Mad Max Beyond Thunderdome 563 Mary Poppins 6879 Maya Deren...Shorts: Meshes in the Afternoon...(Silent 1943) 4882 Nice Girls: films by and about women 3052 Paperhouse 1768 Peggy Sue Got Married 8519 Pleasantville 754 Poltergeist 6898 Poor Little Rich Girl (1917 B&W Silent) 2042 Power of Myth: First Storytellers 2040 Power of Myth: Hero’s Adventure 2044 Power of Myth: Love and the Goddess 2045 -

Florida Waters
Florida A Water Resources Manual from Florida’s Water Management Districts Credits Author Elizabeth D. Purdum Institute of Science and Public Affairs Florida State University Cartographer Peter A. Krafft Institute of Science and Public Affairs Florida State University Graphic Layout and Design Jim Anderson, Florida State University Pati Twardosky, Southwest Florida Water Management District Project Manager Beth Bartos, Southwest Florida Water Management District Project Coordinators Sally McPherson, South Florida Water Management District Georgann Penson, Northwest Florida Water Management District Eileen Tramontana, St. Johns River Water Management District For more information or to request additional copies, contact the following water management districts: Northwest Florida Water Management District 850-539-5999 www.state.fl.us/nwfwmd St. Johns River Water Management District 800-451-7106 www.sjrwmd.com South Florida Water Management District 800-432-2045 www.sfwmd.gov Southwest Florida Water Management District 800-423-1476 www.WaterMatters.org Suwannee River Water Management District 800-226-1066 www.mysuwanneeriver.com April 2002 The water management districts do not discriminate upon the basis of any individual’s disability status. Anyone requiring reasonable accommodation under the ADA should contact the Communications and Community Affairs Department of the Southwest Florida Water Management District at (352) 796-7211 or 1-800-423-1476 (Florida only), extension 4757; TDD only 1-800-231-6103 (Florida only). Contents CHAPTER 1 -

Beasts of Tarzan by Edgar Rice Burroughs
The Beasts of Tarzan by Edgar Rice Burroughs To Joan Burroughs CONTENTS CHAPTER PAGE 1 Kidnapped . 1 2 Marooned . 9 3 Beasts at Bay . 18 4 Sheeta . 28 5 Mugambi . 37 6 A Hideous Crew . 46 7 Betrayed . 55 8 The Dance of Death . 64 9 Chivalry or Villainy . 73 10 The Swede . 82 11 Tambudza . 90 12 A Black Scoundrel . 98 13 Escape . 107 14 Alone in the Jungle . 115 15 Down the Ugambi . 123 16 In the Darkness of the Night . 132 17 On the Deck of the "Kincaid" . 140 18 Paulvitch Plots Revenge . 147 19 The Last of the "Kincaid" . 158 20 Jungle Island Again . 162 21 The Law of the Jungle . 172 Chapter 1 Kidnapped "The entire affair is shrouded in mystery," said D'Arnot. "I have it on the best of authority that neither the police nor the special agents of the general staff have the faintest conception of how it was accomplished. All they know, all that anyone knows, is that Nikolas Rokoff has escaped." John Clayton, Lord Greystoke--he who had been "Tarzan of the Apes"-- sat in silence in the apartments of his friend, Lieutenant Paul D'Arnot, in Paris, gazing meditatively at the toe of his immaculate boot. His mind revolved many memories, recalled by the escape of his arch-enemy from the French military prison to which he had been sentenced for life upon the testimony of the ape-man. He thought of the lengths to which Rokoff had once gone to compass his death, and he realized that what the man had already done would doubtless be as nothing by comparison with what he would wish and plot to do now that he was again free. -

2001 Annual Report
wdwCovers 12/18/01 5:17 PM Page 1 The Company ANNUAL REPORT 2001 wdwCovers 12/18/01 5:17 PM Page 2 Reveta F. Bowers John E. Bryson Roy E. Disney Michael D. Eisner Judith L. Estrin Stanley P. Gold Robert A. Iger Monica C. Lozano George J. Mitchell Thomas S. Murphy Leo J. O’Donovan, S.J. Sidney Poitier Robert A.M. Stern Andrea L. Van de Kamp Raymond L. Watson Gary L. Wilson 20210F01_P01.09_v2 12/18/01 5:19 PM Page 1 The Walt Disney Company and Subsidiaries CONTENT LISTING Financial Highlights 1 Management’s Discussion and Analysis 49 Letter to Shareholders 2 Consolidated Statements of Income 60 Financial Review 10 Consolidated Balance Sheets 61 DisneyHand 14 Consolidated Statements of Cash Flows 62 Parks and Resorts 18 Consolidated Statements of Stockholders’ Equity 63 Walt Disney Imagineering 26 Notes to Consolidated Financial Statements 64 Studio Entertainment 28 Quarterly Financial Summary 77 Media Networks 36 Selected Financial Data 78 Broadcast Networks 37 Management’s Responsibility of Financial Statements 79 Cable Networks 38 Report of Independent Accountants 79 Consumer Products 44 Board of Directors and Corporate Executive Officers 80 Walt Disney International 48 FINANCIAL HIGHLIGHTS (In millions, except per share data) 2001 2000 Revenues(1) $25,256 $25,356 Segment operating income(1) 4,038 4,124 Diluted earnings per share before the cumulative effect of accounting changes, excluding restructuring and impairment charges and gain on the sale of businesses(1) 0.72 0.72 Cash flow from operations 3,048 3,755 Borrowings 9,769 9,461 Stockholders’ equity 22,672 24,100 (1) Pro forma revenues, segment operating income and earnings per share reflect the sale of Fairchild Publications, the acquisition of Infoseek, the conversion of Internet Group common stock into Disney common stock and the closure of the GO.com portal business as if these events and the adoption of SOP 00-2 had occurred at the beginning of fiscal 2000, eliminating the one-time impact of those events. -
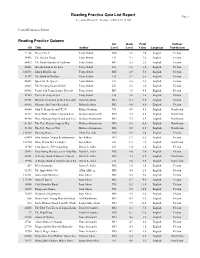
Reading Practice Quiz List Report Page 1 Accelerated Reader®: Tuesday, 11/06/12, 09:18 AM
Reading Practice Quiz List Report Page 1 Accelerated Reader®: Tuesday, 11/06/12, 09:18 AM Central Elementary School Reading Practice Quizzes Int. Book Point Fiction/ ID Title Author Level Level Value Language Non-fiction 71266 Dream Thief Tony Abbott MG 3.8 2.0 English Fiction 54492 The Golden Wasp Tony Abbott LG 3.1 1.0 English Fiction 54493 The Hawk Bandits of Tarkoom Tony Abbott MG 3.5 2.0 English Fiction 54481 Into the Land of the Lost Tony Abbott LG 3.3 1.0 English Fiction 145078 Lunch-Box Dream Tony Abbott MG 4.9 5.0 English Fiction 54497 The Mask of Maliban Tony Abbott LG 3.7 2.0 English Fiction 54487 Quest for the Queen Tony Abbott LG 3.3 1.0 English Fiction 39832 The Sleeping Giant of Goll Tony Abbott LG 3.0 1.0 English Fiction 60582 Trapped in Transylvania: Dracula Tony Abbott MG 3.8 4.0 English Fiction 54503 Under the Serpent Sea Tony Abbott LG 3.6 2.0 English Fiction 89001 Mission: In Search of the Time and SpaceDeborah Machine Abela MG 5.1 5.0 English Fiction 89002 Mission: Spy Force Revealed Deborah Abela MG 6.0 8.0 English Fiction 66660 John F. Kennedy and PT109 Philip Abraham UG 4.9 0.5 English Nonfiction 44342 Main Battle Tanks (Land and Sea) Melissa Abramovitz MG 5.3 0.5 English Nonfiction 44360 Mine Hunting Ships (Land and Sea) Melissa Abramovitz MG 5.3 0.5 English Nonfiction 51153 The U.S. Marine Corps at War Melissa Abramovitz MG 5.9 0.5 English Nonfiction 51154 The U.S.