Curriculum Vitae Dr
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

from Within and Without the University
February 15-16, 2013 : From Within and Without the University Sponsored by Martha B. Reynolds Endowment in Writing, Rhetoric and Digital Media College of Arts & Sciences Office of the Vice President for Research SCHEDULE FEBRUARY 15, 2013 Patterson Office Tower 18th floor 8:00-8:10 Mark Kornbluh, Dean of the College of Arts & Sciences 8:10-8:20 Welcome 8:30 – 9:30 am Panel A New Directions in Rhetorical Listening “Echolocating Rhetorical Listening,” Kyle Jensen, University of North Texas “Listening with Robots,” Jodie Nicotra, University of Idaho “Becoming Rhetorical Listening,” Julie Jung, Illinois State University Panel B Publishing Networks “Networked Humanities Scholarship, or the Life of Kairos” Cheryl Ball, Illinois State University and Douglas Eyman, George Mason University “Public Philosophy Journal” Chris Long and Mark Fisher, The Pennsylvania State University 9:40 – 10:40 am Panel A Religion Networks “E-vangelism and the Rhetoric of Digital Bridge Building, ” Amber Stamper, University of Kentucky “Networked Kairos: The Persuasive Power of Fundamentalist Rhetorics,” Naomi Clark, University of Missouri “Digitizing Heaven: Iconic (Re)Production in Digital Networks,” Amy Anderson, University of Kentucky Panel B Knowledge Networks “Reading in Slow Motion: Thinking With the Network” Jillian Sayre and James J. Brown, Jr., University of Wisconsin “Africa in the Picture: Curated Collections of Images of Africa for Interdisciplinary Use,” Monicá Blackmun Visona and Lesley Chapman, University of Kentucky 10:50-11:50 am Panel A Personal -

Auburn Vs Clemson (10/27/1962)
Clemson University TigerPrints Football Programs Programs 1962 Auburn vs Clemson (10/27/1962) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Auburn vs Clemson (10/27/1962)" (1962). Football Programs. 56. https://tigerprints.clemson.edu/fball_prgms/56 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. CLEMSON MEMORIAL 5TA0IUM-2RM. CLEMSON OCT -27/ AUBURN OFFICIAL PR.OO'RAM 50<t= 7 Thru-Liners Daily FOR SAFETY - CONVENIENCE As Follows: Via Atlanta. Ga. To Houston Texas Via Atlanta to COMFORT AND ECONOMY Jackson, Miss. Via Atlanta to Tallahassee, Fla. Via Atlanta to Dallas, Texas Via Atlanta to Wichita Falls. Texas Via Atlanta to Texarkana, Texas Via Atlanta to New Orleans, La. Three Thru -Lines Daily to Norfolk, Va. & Two Trips Daily to Columbia and Myrtle Beach & Seven Thru Trips AIR- SUSPENSION Daily to Charlotte, N. C. (Thru-Liners) Six Trips Daily to TRAILWAYS COACHES New York City (Three Thru-Liners) Three Thru-Liners Daily To Cleveland, Ohio* fe You board and leave your . -

Georgia Tech Vs Clemson (11/12/1994)
Clemson University TigerPrints Football Programs Programs 1994 Georgia Tech vs Clemson (11/12/1994) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Georgia Tech vs Clemson (11/12/1994)" (1994). Football Programs. 232. https://tigerprints.clemson.edu/fball_prgms/232 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. For nearly half a century, your global partner for textile technology • Alexco: Fabric take-ups, let-offs and inspection frames •Jenkins: Waste briquetting press, circular fans • Beltran: Pollution and smoke abatement equipment •Juwon: Sock knitting machinery •Dernier: Universal weaving machine, air jet or rapier •Knotex: Warp tying and drawing-in systems •Ducker: Dryers and wrinkle-free curing ovens •Lemaire: Transfer printing for fabric and -

Duke Vs Clemson (11/7/1959)
Clemson University TigerPrints Football Programs Programs 1959 Duke vs Clemson (11/7/1959) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Duke vs Clemson (11/7/1959)" (1959). Football Programs. 41. https://tigerprints.clemson.edu/fball_prgms/41 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. cmsou DUKE Cleiti^oit Memorial Stadium -*-N0V.7/ lose -^OFFICIAL PRO&^m-k 50^ SACO-LOWELL SETTING THE PACE FOR THE TEXTILE INDUSTRY THROUGH RESEARCH & DEVELOPME D*l^ ,.,,,»MHiiiiiiiPWfyii«iWiiMiilM^^ Research and Development Center Clemson, S. C. Manufacturers of a complete line of yarn preparatory machin- ery for opening through spinning for processing cottons, worsteds and synthetics, and the revolutionary Fleissner Dryer. SACO-LOWELLSACO-LOWELLTEXTILE MACHINERYSHOPSDIVISION fitcutivt < Solri Offrcci — lASlIV, $ C S C ones Selci Officei — ATLANTA. CA , SOSTON, MASS, CHAdOTTE « CIEENSBORO. N C & GREENVILK TIMELY because they care how it fits and how it looks . -

Auburn Vs Clemson (10/10/1970)
Clemson University TigerPrints Football Programs Programs 1970 Auburn vs Clemson (10/10/1970) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Auburn vs Clemson (10/10/1970)" (1970). Football Programs. 90. https://tigerprints.clemson.edu/fball_prgms/90 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. Stevens-Urica® No-Iron Sheets They don't just make beds. They make bedrooms. Official Program Published By ATHLETIC DEPARTMENT CLEMSON UNIVERSITY Edited By BOB BRADLEY Director of Sports Information Assisted By JERRY ARP Ass't. Sports Information Director Represented for National Advertising By SPENCER MARKETING SERVICES 370 Lexington Avenue New York. New York 10017 Photography by Jim Burns, Charles Haralson, Tom Shockley, Hal Smith, and Bill Osteen of Clemson; Jim Laughead and Jim Bradley of Dallas, Texas IMPORTANT EMERGENCIES: A first aid station is located LOST & FOUND: If any article is lost or found, under Section A on South side of Stadium. -

Patricia Ann Browne-Ferrigno
Tricia Browne-Ferrigno, PhD Department of Educational Leadership Studies University of Kentucky, 111 Dickey Hall, Lexington, KY 40506-0017 Phone: (859) 257-5504 Email: [email protected] ACADEMIC DEGREES PhD University of Colorado Denver, 2001 Educational Leadership and Innovation/Educational Policy and Administration MA University of South Florida, 1996 Special Education/Gifted BA Florida State University, 1968 Mathematics Education/Secondary (Grades 6-12) EMPLOYMENT AND WORK EXPERIENCES 2013-Present Professor, Educational Leadership Studies, University of Kentucky 2011-2013 Associate Professor/Teacher Leadership Program Chair 2007-2011 Associate Professor/Director of Graduate Studies 2001-2007 Assistant Professor/Director of Externally Funded Projects Coordinate graduate-level preparation program for teacher leaders and two graduate certificate programs. Conducted case study on doctoral program redesign as member of national research team for the Carnegie Project on the Education Doctorate (CPED). Served four-year term as department’s Director of Graduate Studies responsible for student recruitment, admissions, advising, and examination and for coordination of program development, implementation, and evaluation. Served as faculty member and dissertation advisor for CPED-affiliated EdD cohort program to prepare community and technical college leaders. Conducted case studies of two university-based preparation programs as senior researcher on national study to assess influence of district-university collaboration on principal preparation, supported by grant from The Wallace Foundation. Directed multi-partner project to develop broad-based instructional leadership teams in high schools, supported by state grant from US Department of Education Improving Teacher Quality Program. Directed three-year professional development project for principals and administrator-credentialed teachers in rural Appalachia district, supported by grant from US Department of Education School Leadership Program and featured in Innovative Pathways to School Leadership. -

Virginia Tech Vs Clemson (10/7/1978)
Clemson University TigerPrints Football Programs Programs 1978 Virginia Tech vs Clemson (10/7/1978) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Virginia Tech vs Clemson (10/7/1978)" (1978). Football Programs. 133. https://tigerprints.clemson.edu/fball_prgms/133 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. Eastern Distribution is people who Eastern Distribution Executive Vice President E. Harold Segars, Jr. know how to handle things People who can get anything at all from one place to another on the right timetable, and in perfect condition. Eastern Distribution Office Manager Dianne Moore, Sales Representative Sherry Turner, and Controller Carrol Garrett Yes, the Eastern people on Harold Segars' Greenville, S. C, and Jacksonville, Fla., distribution team get things done, whether they're arranging the same-day movement of something you want out in a hurry, or consolidating loads to save you money through lower rates. -

Georgia Tech Vs Clemson (9/28/1974)
Clemson University TigerPrints Football Programs Programs 1974 Georgia Tech vs Clemson (9/28/1974) Clemson University Follow this and additional works at: https://tigerprints.clemson.edu/fball_prgms Materials in this collection may be protected by copyright law (Title 17, U.S. code). Use of these materials beyond the exceptions provided for in the Fair Use and Educational Use clauses of the U.S. Copyright Law may violate federal law. For additional rights information, please contact Kirstin O'Keefe (kokeefe [at] clemson [dot] edu) For additional information about the collections, please contact the Special Collections and Archives by phone at 864.656.3031 or via email at cuscl [at] clemson [dot] edu Recommended Citation University, Clemson, "Georgia Tech vs Clemson (9/28/1974)" (1974). Football Programs. 110. https://tigerprints.clemson.edu/fball_prgms/110 This Book is brought to you for free and open access by the Programs at TigerPrints. It has been accepted for inclusion in Football Programs by an authorized administrator of TigerPrints. For more information, please contact [email protected]. CLEMS;feN TIGERS THE GEORGIA TECH GAME SEPT. 28, 1974 hOO P.M. ^ CLEMSON MEMORIAL STADIUM $1.00 Ideally situated to save you time and money. When Eastern meets your distribution needs, you have an experienced group working for you in two ideal locations: Greenville, South Carolina, and Jacksonville, Florida. The recent addition of two brand new distribution centers in Imeson Park at Jacksonville gives us total floor space of 1 ,167,000 sq. ft., with more projected. Our matenals handling and warehouse maintenance equipment is the finest. Our personnel hand picked. -
Express Yourself
Express yourself Find the right card for you When you’re opening your new eligible checking account, consider getting a custom debit card that says something about you. As an alumnus, sports fan, professional or Charitable Causes supporter of a special cause, you have a choice of debit cards that bear the name of an organization you care about. Every account is offered and serviced by Bank of America. Colleges & Universities Click on any of these images to find the right card for you. Request yours by calling 800.432.1000 or visiting a Bank of America Military Themes financial center. Professional Sports & Recreation The banking you do can help generate contributions to the cause you care about —at no additional expense Charitable Causes to you. You get the same great banking product and service; the organization gets the support they deserve. Defenders of Wildlife Golden Key International National Trust for National Wildlife Federation ID# 23328 Honour Society Historic Preservation ID# 10358 ID# 11397 ID# 11353 Susan G. Komen The Nature Conservancy World Wildlife Fund ID# 62888 ID# 23032 ID# 62812 ❮BACK HOME NEXT❯ Bank of America has partnered with dozens of colleges and universities to offer alumni a banking program that can help generate contributions to their alma maters. Show your school spirit with a debit card Colleges & Universities emblazoned with your school’s name and logo! Alumni Association of the Arkansas Alumni Association Brown Alumni Association Cal Alumni Association University of Virginia ID# 19449 ID# 23303 ID# 23312 ID# 63048 Clemson University Columbia Alumni Association Florida International Fordham University ID# 15553 ID# 11561 University ID# 13873 ID# 13308 General Alumni Association Georgetown University Indiana University Iowa State University UNC Chapel Hill Alumni Association Alumni Association Alumni Association ID# 16559 ID# 10602 ID# 12595 ID# 15731 ❮BACK HOME NEXT❯ Bank of America has partnered with dozens of colleges and universities to offer alumni a banking program that can help generate contributions to their alma maters. -
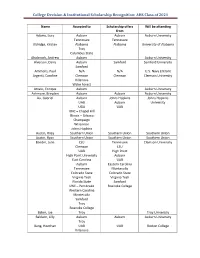
College Decision & Institutional Scholarship Recognition: AHS Class of 2021
College Decision & Institutional Scholarship Recognition: AHS Class of 2021 Name Accepted to Scholarship offers Will be attending from Adams, Lucy Auburn Auburn Auburn University Tennessee Tennessee Aldridge, Kristen Alabama Alabama University of Alabama Troy Columbus State Alsobrook, Andrew Auburn Auburn University Alverson, Davis Auburn Samford Samford University Samford Ammons, Paul N/A N/A U.S. Navy Entrant Argenti, Caroline Clemson Clemson Clemson University Villanova Wake Forest Arvelo, Enrique Auburn Auburn University Ashmore, Brayden Auburn Auburn Auburn University Au, Gabriel Auburn Johns Hopkins Johns Hopkins UAB Auburn University UGA UAB UNC – Chapel Hill Illinois – Urbana Champaign Wisconsin Johns Hopkins Austin, Riley Southern Union Southern Union Southern Union Austin, Ryan Southern Union Southern Union Southern Union Baeder, Julie LSU Tennessee Clemson University Clemson LSU UAB High Point High Point University Auburn East Carolina UAB Auburn Eastern Carolina Tennessee Montevallo Colorado State Colorado State Virginia Tech Virginia Tech Florida State Samford UNC – Pembroke Roanoke College Western Carolina Montevallo Samford Troy Roanoke College Baker, Jae Troy Troy University Baldwin, Lilly Auburn Auburn Auburn University Troy Bang, Heechan UAB UAB Boston College Villanova College Decision & Institutional Scholarship Recognition: AHS Class of 2021 Boston College Bang, Yeseul Auburn Auburn Auburn University Alabama Alabama Barjis, Ariabel Auburn IUPUI Indiana University – Indiana University – Auburn Purdue University Purdue -

Graduation Rates at Auburn and SREB Peers
Graduation Rates at Auburn and SREB Peers A comparison of recent graduation rates at Auburn University and its SREB peer institutions places Auburn at the median and second quartile for 2004-2006 reporting years (see Fig 1 for the distribution of completion rates; see Table 1 for the list of peer institutions). Differences in graduation rate by gender and ethnicity appear across the SREB peer group, including Auburn. Women are more likely than men to graduate (see Fig 2); again, a comparison with peer institutions places Auburn at the median and second quartile. Black students are less likely than students generally to graduate. Auburn’s graduation rates for black students are in the second quartile in 2004 and 2005 and in the lowest quartile in 2006 (see Fig 3). Across the SREB, Asian students are more likely to graduate than students generally. However, at Auburn graduation rates for Asian students are lower than for students generally and place in the lowest quartile for 2004 and 2005 and in the second quartile for 2006 (see Fig 4). This brief comparison demonstrates that Auburn does not outperform the middle point for its peers and calls for a more detailed analysis of six-year graduation rates at Auburn. Fig 1 Graduation rate - Bachelor degree within 6 Fig 2 Graduation rate - Bachelor degree within 6 years, total* years, women* 100 100 90 90 80 80 70 70 69 65 66 67 63 60 62 60 50 50 40 40 30 30 2004 2005 2006 2004 2005 2006 Fig 3 Graduation rate - Bachelor degree within 6 Fig 4 Graduation rate - Bachelor degree within 6 years, Black, -

Robert Elder Current Cv Template
Robert Elder Assistant Professor of History Baylor University History Department One Bear Place #97306 Waco, TX 76798 Phone: 254-710-6211 (office) robert [email protected] Education 2011 Ph.D., history, Emory University 2005 M.A., history, Clemson University 2003 B.A. summa cum laude, history and English, Clemson University Professional 2018- Assistant Professor of History, Baylor University 2014-2018 Assistant Professor of History, Valparaiso University 2013-2014 Assistant Professor of History, Tabor College 2011-2013 Lilly Postdoctoral Fellow and Lecturer in Humanities and History, Christ College, Valparaiso University Distinctions 2016-2017 University Research Professorship, Valparaiso University 2015 Creative Work and Research Committee Summer Fellowship, Valparaiso Uni- versity 2012 Dissertation nominated by Emory University history department for the 2012 Allan Nevins Prize, o↵ered by the Society of American Historians for the best-written doctoral dissertation on an American subject 2010-2011 Dean’s Teaching Fellowship, Laney Graduate School, Emory University (for proven excellence and potential in teaching) 2009 Lewis P. Jones Research Fellow, South Caroliniana Library, University of South Carolina 2008, 2009 Competitive Research Award, Graduate School of Arts & Sciences, Emory University 2007, 2008 Visiting Research Fellowship, Institute for Southern Studies, University of South Carolina 2005 South Carolina Colonial Dames Award for outstanding graduate student in American History, Clemson University Publications Books John C.