Advance Experimental Taxonomy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Natural Heritage Program List of Rare Plant Species of North Carolina 2016
Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Revised February 24, 2017 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org C ur Alleghany rit Ashe Northampton Gates C uc Surry am k Stokes P d Rockingham Caswell Person Vance Warren a e P s n Hertford e qu Chowan r Granville q ot ui a Mountains Watauga Halifax m nk an Wilkes Yadkin s Mitchell Avery Forsyth Orange Guilford Franklin Bertie Alamance Durham Nash Yancey Alexander Madison Caldwell Davie Edgecombe Washington Tyrrell Iredell Martin Dare Burke Davidson Wake McDowell Randolph Chatham Wilson Buncombe Catawba Rowan Beaufort Haywood Pitt Swain Hyde Lee Lincoln Greene Rutherford Johnston Graham Henderson Jackson Cabarrus Montgomery Harnett Cleveland Wayne Polk Gaston Stanly Cherokee Macon Transylvania Lenoir Mecklenburg Moore Clay Pamlico Hoke Union d Cumberland Jones Anson on Sampson hm Duplin ic Craven Piedmont R nd tla Onslow Carteret co S Robeson Bladen Pender Sandhills Columbus New Hanover Tidewater Coastal Plain Brunswick THE COUNTIES AND PHYSIOGRAPHIC PROVINCES OF NORTH CAROLINA Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org This list is dynamic and is revised frequently as new data become available. New species are added to the list, and others are dropped from the list as appropriate. -

Seed Ecology Iii
SEED ECOLOGY III The Third International Society for Seed Science Meeting on Seeds and the Environment “Seeds and Change” Conference Proceedings June 20 to June 24, 2010 Salt Lake City, Utah, USA Editors: R. Pendleton, S. Meyer, B. Schultz Proceedings of the Seed Ecology III Conference Preface Extended abstracts included in this proceedings will be made available online. Enquiries and requests for hardcopies of this volume should be sent to: Dr. Rosemary Pendleton USFS Rocky Mountain Research Station Albuquerque Forestry Sciences Laboratory 333 Broadway SE Suite 115 Albuquerque, New Mexico, USA 87102-3497 The extended abstracts in this proceedings were edited for clarity. Seed Ecology III logo designed by Bitsy Schultz. i June 2010, Salt Lake City, Utah Proceedings of the Seed Ecology III Conference Table of Contents Germination Ecology of Dry Sandy Grassland Species along a pH-Gradient Simulated by Different Aluminium Concentrations.....................................................................................................................1 M Abedi, M Bartelheimer, Ralph Krall and Peter Poschlod Induction and Release of Secondary Dormancy under Field Conditions in Bromus tectorum.......................2 PS Allen, SE Meyer, and K Foote Seedling Production for Purposes of Biodiversity Restoration in the Brazilian Cerrado Region Can Be Greatly Enhanced by Seed Pretreatments Derived from Seed Technology......................................................4 S Anese, GCM Soares, ACB Matos, DAB Pinto, EAA da Silva, and HWM Hilhorst -

Genetic Diversity and Population Structure of Haloxylon Salicornicum Moq
RESEARCH ARTICLE Genetic diversity and population structure of Haloxylon salicornicum moq. in Kuwait by ISSR markers 1 1 1 1 2 Fadila Al SalameenID *, Nazima HabibiID , Vinod Kumar , Sami Al Amad , Jamal Dashti , Lina Talebi3, Bashayer Al Doaij1 1 Biotechnology Program, Environment and Life Sciences Research Centre, Kuwait Institute for Scientific Research, Shuwaikh, Kuwait, 2 Desert, Agriculture & Ecosystems Program, Environment and Life Sciences a1111111111 Research Centre, Kuwait Institute for Scientific Research, Shuwaikh, Kuwait, 3 Environment Pollution and a1111111111 Climate Change Program, Environment and Life Sciences Research Centre, Kuwait Institute for Scientific a1111111111 Research, Shuwaikh, Kuwait a1111111111 * [email protected] a1111111111 Abstract OPEN ACCESS Haloxylon salicornicum moq. Bunge ex Boiss (Rimth) is one of the native plants of Kuwait, extensively depleting through the anthropogenic activities. It is important to conserve Halox- Citation: Al Salameen F, Habibi N, Kumar V, Al Amad S, Dashti J, Talebi L, et al. (2018) Genetic ylon community in Kuwait as it can tolerate extreme adverse conditions of drought and salin- diversity and population structure of Haloxylon ity to be potentially used in the desert and urban revegetation and greenery national salicornicum moq. in Kuwait by ISSR markers. programs. Therefore, a set of 16 inter simple sequence repeat (ISSR) markers were used to PLoS ONE 13(11): e0207369. https://doi.org/ 10.1371/journal.pone.0207369 assess genetic diversity and population structure of 108 genotypes from six locations in Kuwait. The ISSR primers produced 195 unambiguous and reproducible bands out of which Editor: Ruslan Kalendar, University of Helsinki, FINLAND 167 bands were polymorphic (86.5%) with a mean PIC value of 0.31. -

Vegetation Succession Along New Roads at Soqotra Island (Yemen): Effects of Invasive Plant Species and Utilization of Selected N
10.2478/jlecol-2014-0003 Journal of Landscape Ecology (2013), Vol: 6 / No. 3. VEGETATION SUCCESSION ALONG NEW ROADS AT SOQOTRA ISLAND (YEMEN): EFFECTS OF INVASIVE PLANT SPECIES AND UTILIZATION OF SELECTED NATIVE PLANT RESISTENCE AGAINST DISTURBANCE PETR MADĚRA1, PAVEL KOVÁŘ2, JAROSLAV VOJTA2, DANIEL VOLAŘÍK1, LUBOŠ ÚRADNÍČEK1, ALENA SALAŠOVÁ3, JAROSLAV KOBLÍŽEK1 & PETR JELÍNEK1 1Mendel University in Brno, Faculty of Forestry and Wood Technology, Department of the Forest Botany, Dendrology and Geobiocoenology, Zemědělská 1/1665, 613 00 Brno 2Charles University in Prague, Faculty of Science, Department of Botany, Benátská 2, 128 01 Prague 3Mendel University in Brno, Faculty of Horticulture, Department of Landscape Planning, Valtická 337, 691 44 Lednice Received: 13th November 2013, Accepted: 17th December 2013 ABSTRACT The paved (tarmac) roads had been constructed on Soqotra island over the last 15 years. The vegetation along the roads was disturbed and the erosion started immediately after the disturbance caused by the road construction. Our assumption is that biotechnical measurements should prevent the problems caused by erosion and improve stabilization of road edges. The knowledge of plant species which are able to grow in unfavourable conditions along the roads is important for correct selection of plants used for outplanting. The vegetation succession was observed using phytosociological relevés as a tool of recording and mapping assambblages of plants species along the roads as new linear structures in the landscape. Data from phytosociological relevés were analysed and the succession was characterised in different altitudes. The results can help us to select group of plants (especially shrubs and trees), which are suitable to be used as stabilizing green mantle in various site conditions and for different purposes (anti-erosional, ornamental, protection against noise or dust, etc.). -

In Vivo Hypoglycemic Effect of Methanolic Fruit Extract of Momordica Charantia L
In vivo hypoglycemic effect of methanolic fruit extract of Momordica charantia L Nkambo W1, *Anyama NG, Onegi B Department of Pharmacy, School of Health Sciences, College of Health Sciences, Makerere University, P.O. Box 7072, Kampala, Uganda Abstract Background: Momordica charantia L. is a medicinal plant commonly used in the management of diabetes mellitus. Objectives: We investigated the blood glucose lowering effect of the methanolic fruit extract of the Ugandan variety of M. charantia L. in alloxan-induced diabetic albino rats. Methods: 500g of M. charantia powder were macerated in methanol and the extract administered to two groups of alloxan- induced diabetic rats. The first group received 125mg/kg, the second 375mg/kg and a third group 7mg/ kg of metformin. A fourth group received 1ml normal saline. Fasting blood glucose (FBG) levels were measured at 0.5,1,2,3,5,8 and 12 hours and compared using one-way ANOVA. Results: There was an initial rise in FBG for 1 hour after administration of extracts followed by steep reductions. Significant reduction in FBG occurred at 2 hours for 125mg/kg of extract (-3.2%, 313±25.9 to 303±25.0mg/dL, p = 0.049), 375mg/ kg of extract (-3.9%, 356±19.7 to 342±20.3mg/dL, p = 0.001), and metformin (-2.6%, 344±21.7 to 335±21.1mg/dL, p = 0.003) when compared to normal saline. The maximum percentage reduction in FBG by both extracts occurred between 3 and 12 hours post dose. Conclusions: The methanolic fruit extract of M. charantia exhibits dose dependent hypoglycaemic activity in vivo. -

Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1
Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1 Authors: Jiang, Wei, He, Hua-Jie, Lu, Lu, Burgess, Kevin S., Wang, Hong, et. al. Source: Annals of the Missouri Botanical Garden, 104(2) : 171-229 Published By: Missouri Botanical Garden Press URL: https://doi.org/10.3417/2019337 BioOne Complete (complete.BioOne.org) is a full-text database of 200 subscribed and open-access titles in the biological, ecological, and environmental sciences published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Complete website, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/terms-of-use. Usage of BioOne Complete content is strictly limited to personal, educational, and non - commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Downloaded From: https://bioone.org/journals/Annals-of-the-Missouri-Botanical-Garden on 01 Apr 2020 Terms of Use: https://bioone.org/terms-of-use Access provided by Kunming Institute of Botany, CAS Volume 104 Annals Number 2 of the R 2019 Missouri Botanical Garden EVOLUTION OF ANGIOSPERM Wei Jiang,2,3,7 Hua-Jie He,4,7 Lu Lu,2,5 POLLEN. 7. NITROGEN-FIXING Kevin S. Burgess,6 Hong Wang,2* and 2,4 CLADE1 De-Zhu Li * ABSTRACT Nitrogen-fixing symbiosis in root nodules is known in only 10 families, which are distributed among a clade of four orders and delimited as the nitrogen-fixing clade. -

Natural Heritage Program List of Rare Plant Species of North Carolina 2012
Natural Heritage Program List of Rare Plant Species of North Carolina 2012 Edited by Laura E. Gadd, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program Office of Conservation, Planning, and Community Affairs N.C. Department of Environment and Natural Resources 1601 MSC, Raleigh, NC 27699-1601 Natural Heritage Program List of Rare Plant Species of North Carolina 2012 Edited by Laura E. Gadd, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program Office of Conservation, Planning, and Community Affairs N.C. Department of Environment and Natural Resources 1601 MSC, Raleigh, NC 27699-1601 www.ncnhp.org NATURAL HERITAGE PROGRAM LIST OF THE RARE PLANTS OF NORTH CAROLINA 2012 Edition Edited by Laura E. Gadd, Botanist and John Finnegan, Information Systems Manager North Carolina Natural Heritage Program, Office of Conservation, Planning, and Community Affairs Department of Environment and Natural Resources, 1601 MSC, Raleigh, NC 27699-1601 www.ncnhp.org Table of Contents LIST FORMAT ......................................................................................................................................................................... 3 NORTH CAROLINA RARE PLANT LIST ......................................................................................................................... 10 NORTH CAROLINA PLANT WATCH LIST ..................................................................................................................... 71 Watch Category -

Towards an Updated Checklist of the Libyan Flora
Towards an updated checklist of the Libyan flora Article Published Version Creative Commons: Attribution 3.0 (CC-BY) Open access Gawhari, A. M. H., Jury, S. L. and Culham, A. (2018) Towards an updated checklist of the Libyan flora. Phytotaxa, 338 (1). pp. 1-16. ISSN 1179-3155 doi: https://doi.org/10.11646/phytotaxa.338.1.1 Available at http://centaur.reading.ac.uk/76559/ It is advisable to refer to the publisher’s version if you intend to cite from the work. See Guidance on citing . Published version at: http://dx.doi.org/10.11646/phytotaxa.338.1.1 Identification Number/DOI: https://doi.org/10.11646/phytotaxa.338.1.1 <https://doi.org/10.11646/phytotaxa.338.1.1> Publisher: Magnolia Press All outputs in CentAUR are protected by Intellectual Property Rights law, including copyright law. Copyright and IPR is retained by the creators or other copyright holders. Terms and conditions for use of this material are defined in the End User Agreement . www.reading.ac.uk/centaur CentAUR Central Archive at the University of Reading Reading’s research outputs online Phytotaxa 338 (1): 001–016 ISSN 1179-3155 (print edition) http://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2018 Magnolia Press Article ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.338.1.1 Towards an updated checklist of the Libyan flora AHMED M. H. GAWHARI1, 2, STEPHEN L. JURY 2 & ALASTAIR CULHAM 2 1 Botany Department, Cyrenaica Herbarium, Faculty of Sciences, University of Benghazi, Benghazi, Libya E-mail: [email protected] 2 University of Reading Herbarium, The Harborne Building, School of Biological Sciences, University of Reading, Whiteknights, Read- ing, RG6 6AS, U.K. -

Taxonomic Significance of Spermoderm Pattern in Cucurbitaceae M. Ajmal
Bangladesh J. Plant Taxon. 20(1): 61-65, 2013 (June) © 2013 Bangladesh Association of Plant Taxonomists TAXONOMIC SIGNIFICANCE OF SPERMODERM PATTERN IN CUCURBITACEAE 1 2 3 M. AJMAL ALI , FAHAD M.A. AL-HEMAID, ARUN K. PANDEY AND JOONGKU LEE Department of Botany and Microbiology, College of Science, King Saud University, Riyadh-11451, Saudi Arabia. Keywords: Cucurbitaceae; SEM; Spermoderm; Testa. Abstract Studies on spermoderm using scanning electron microscope (SEM) were undertaken in 12 taxa under 11 genera of the family Cucurbitaceae sampled from India, China and Korea. The spermoderm pattern in the studied taxa varies from rugulate, reticulate to colliculate type. The spermoderm shows rugulate type in Benincasa hispida and Sicyos angulatus; reticulate type in Citrullus colocynthis, Cucumis melo var. agrestis, Diplocyclos palmatus, Hemsleya longivillosa, Luffa echinata, Momordica charantia, M. cymbalaria, Schizopepon bryoniifolius, and Trichosanthes cucumerina; and colliculate type in Gynostemma laxiflorum. The present study clearly reveals that the testa features greatly varies across the genera which can be used as micromorphological markers for identification as well as character states for deducing relationship of the taxa within the family. Introduction Spermoderm refers to the pattern present on the seed coat of mature seeds. Seed characteristic, particularly exomorphic features as revealed by scanning electron microscopy, have been used by many workers in resolving taxonomic problems (Koul et al., 2000; Pandey and Ali, 2006) and evolutionary relationships (Kumar et al., 1999; Segarra and Mateu, 2001). Cucurbitaceae, with c. 800 species under 130 genera (Schaefer and Renner, 2011) are among the economically most important plant families (Kirtikar and Basu, 1975; Chakravarty, 1982; Ali and Pandey, 2006). -

Efficacy and Host Specificity Compared Between Two Populations of The
Biological Control 65 (2013) 53–62 Contents lists available at SciVerse ScienceDirect Biological Control journal homepage: www.elsevier.com/locate/ybcon Efficacy and host specificity compared between two populations of the psyllid Aphalara itadori, candidates for biological control of invasive knotweeds in North America ⇑ Fritzi Grevstad a, , Richard Shaw b, Robert Bourchier c, Paolo Sanguankeo d, Ghislaine Cortat e, Richard C. Reardon f a Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR 97331, USA b CABI, Bakeham Lane, Egham, Surrey TW20 9TY, United Kingdom c Agriculture and AgriFood Canada-Lethbridge Research Centre, Lethbridge, AB, Canada T1J 4B1 d Olympic Natural Resources Center, University of Washington, Forks, WA 98331, USA e CABI, CH 2800 Delemont, Switzerland f USDA Forest Service, Forest Health Technology Enterprise Team, Morgantown, WV 26505, USA highlights graphical abstract " Two populations of the psyllid Aphalara itadori are effective at reducing knotweed growth and biomass. " The two populations differ in their performance among different knotweed species. " Development of A. itadori occurred infrequently on several non-target plant species. " The psyllid exhibited non-preference and an inability to persist on non- target plants. article info abstract Article history: Invasive knotweeds are large perennial herbs in the Polygonaceae in the genus Fallopia that are native to Received 2 February 2012 Asia and invasive in North America. They include Fallopia japonica (Japanese knotweed), F. sachalinensis Accepted 4 January 2013 (giant knotweed), and a hybrid species F. x bohemica (Bohemian knotweed). Widespread throughout Available online 12 January 2013 the continent and difficult to control by mechanical or chemical methods, these plants are good targets for classical biological control. -
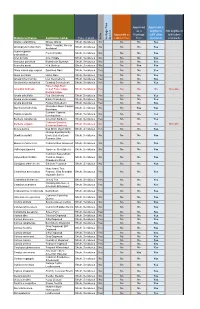
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Genetic Diversity and Evolution in Lactuca L. (Asteraceae)
Genetic diversity and evolution in Lactuca L. (Asteraceae) from phylogeny to molecular breeding Zhen Wei Thesis committee Promotor Prof. Dr M.E. Schranz Professor of Biosystematics Wageningen University Other members Prof. Dr P.C. Struik, Wageningen University Dr N. Kilian, Free University of Berlin, Germany Dr R. van Treuren, Wageningen University Dr M.J.W. Jeuken, Wageningen University This research was conducted under the auspices of the Graduate School of Experimental Plant Sciences. Genetic diversity and evolution in Lactuca L. (Asteraceae) from phylogeny to molecular breeding Zhen Wei Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr A.P.J. Mol, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Monday 25 January 2016 at 1.30 p.m. in the Aula. Zhen Wei Genetic diversity and evolution in Lactuca L. (Asteraceae) - from phylogeny to molecular breeding, 210 pages. PhD thesis, Wageningen University, Wageningen, NL (2016) With references, with summary in Dutch and English ISBN 978-94-6257-614-8 Contents Chapter 1 General introduction 7 Chapter 2 Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons* 31 Chapter 3 Phylogenetic analysis of Lactuca L. and closely related genera (Asteraceae), using complete chloroplast genomes and nuclear rDNA sequences 99 Chapter 4 A mixed model QTL analysis for salt tolerance in