Parallel I/O on Compressed Data Files: Semantics, Algorithms, and Performance Evaluation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Basic Principles of Data Compression
The Basic Principles of Data Compression Author: Conrad Chung, 2BrightSparks Introduction Internet users who download or upload files from/to the web, or use email to send or receive attachments will most likely have encountered files in compressed format. In this topic we will cover how compression works, the advantages and disadvantages of compression, as well as types of compression. What is Compression? Compression is the process of encoding data more efficiently to achieve a reduction in file size. One type of compression available is referred to as lossless compression. This means the compressed file will be restored exactly to its original state with no loss of data during the decompression process. This is essential to data compression as the file would be corrupted and unusable should data be lost. Another compression category which will not be covered in this article is “lossy” compression often used in multimedia files for music and images and where data is discarded. Lossless compression algorithms use statistic modeling techniques to reduce repetitive information in a file. Some of the methods may include removal of spacing characters, representing a string of repeated characters with a single character or replacing recurring characters with smaller bit sequences. Advantages/Disadvantages of Compression Compression of files offer many advantages. When compressed, the quantity of bits used to store the information is reduced. Files that are smaller in size will result in shorter transmission times when they are transferred on the Internet. Compressed files also take up less storage space. File compression can zip up several small files into a single file for more convenient email transmission. -

Encryption Introduction to Using 7-Zip
IT Services Training Guide Encryption Introduction to using 7-Zip It Services Training Team The University of Manchester email: [email protected] www.itservices.manchester.ac.uk/trainingcourses/coursesforstaff Version: 5.3 Training Guide Introduction to Using 7-Zip Page 2 IT Services Training Introduction to Using 7-Zip Table of Contents Contents Introduction ......................................................................................................................... 4 Compress/encrypt individual files ....................................................................................... 5 Email compressed/encrypted files ....................................................................................... 8 Decrypt an encrypted file ..................................................................................................... 9 Create a self-extracting encrypted file .............................................................................. 10 Decrypt/un-zip a file .......................................................................................................... 14 APPENDIX A Downloading and installing 7-Zip ................................................................. 15 Help and Further Reference ............................................................................................... 18 Page 3 Training Guide Introduction to Using 7-Zip Introduction 7-Zip is an application that allows you to: Compress a file – for example a file that is 5MB can be compressed to 3MB Secure the -

The Ark Handbook
The Ark Handbook Matt Johnston Henrique Pinto Ragnar Thomsen The Ark Handbook 2 Contents 1 Introduction 5 2 Using Ark 6 2.1 Opening Archives . .6 2.1.1 Archive Operations . .6 2.1.2 Archive Comments . .6 2.2 Working with Files . .7 2.2.1 Editing Files . .7 2.3 Extracting Files . .7 2.3.1 The Extract dialog . .8 2.4 Creating Archives and Adding Files . .8 2.4.1 Compression . .9 2.4.2 Password Protection . .9 2.4.3 Multi-volume Archive . 10 3 Using Ark in the Filemanager 11 4 Advanced Batch Mode 12 5 Credits and License 13 Abstract Ark is an archive manager by KDE. The Ark Handbook Chapter 1 Introduction Ark is a program for viewing, extracting, creating and modifying archives. Ark can handle vari- ous archive formats such as tar, gzip, bzip2, zip, rar, 7zip, xz, rpm, cab, deb, xar and AppImage (support for certain archive formats depends on the appropriate command-line programs being installed). In order to successfully use Ark, you need KDE Frameworks 5. The library libarchive version 3.1 or above is needed to handle most archive types, including tar, compressed tar, rpm, deb and cab archives. To handle other file formats, you need the appropriate command line programs, such as zipinfo, zip, unzip, rar, unrar, 7z, lsar, unar and lrzip. 5 The Ark Handbook Chapter 2 Using Ark 2.1 Opening Archives To open an archive in Ark, choose Open... (Ctrl+O) from the Archive menu. You can also open archive files by dragging and dropping from Dolphin. -

Working with Compressed Archives
Working with compressed archives Archiving a collection of files or folders means creating a single file that groups together those files and directories. Archiving does not manipulate the size of files. They are added to the archive as they are. Compressing a file means shrinking the size of the file. There are software for archiving only, other for compressing only and (as you would expect) other that have the ability to archive and compress. This document will show you how to use a windows application called 7-zip (seven zip) to work with compressed archives. 7-zip integrates in the context menu that pops-up up whenever you right-click your mouse on a selection1. In this how-to we will look in the application interface and how we manipulate archives, their compression and contents. 1. Click the 'start' button and type '7-zip' 2. When the search brings up '7-zip File Manager' (as the best match) press 'Enter' 3. The application will startup and you will be presented with the 7-zip manager interface 4. The program integrates both archiving, de/compression and file-management operations. a. The main area of the window provides a list of the files of the active directory. When not viewing the contents of an archive, the application acts like a file browser window. You can open folders to see their contents by just double-clicking on them b. Above the main area you have an address-like bar showing the name of the active directory (currently c:\ADG.Becom). You can use the 'back' icon on the far left to navigate back from where you are and go one directory up. -

Deduplicating Compressed Contents in Cloud Storage Environment
Deduplicating Compressed Contents in Cloud Storage Environment Zhichao Yan, Hong Jiang Yujuan Tan* Hao Luo University of Texas Arlington Chongqing University University of Nebraska Lincoln [email protected] [email protected] [email protected] [email protected] Corresponding Author Abstract Data compression and deduplication are two common approaches to increasing storage efficiency in the cloud environment. Both users and cloud service providers have economic incentives to compress their data before storing it in the cloud. However, our analysis indicates that compressed packages of different data and differ- ently compressed packages of the same data are usual- ly fundamentally different from one another even when they share a large amount of redundant data. Existing data deduplication systems cannot detect redundant data among them. We propose the X-Ray Dedup approach to extract from these packages the unique metadata, such as the “checksum” and “file length” information, and use it as the compressed file’s content signature to help detect and remove file level data redundancy. X-Ray Dedup is shown by our evaluations to be capable of breaking in the boundaries of compressed packages and significantly Figure 1: A user scenario on cloud storage environment reducing compressed packages’ size requirements, thus further optimizing storage space in the cloud. will generate different compressed data of the same con- tents that render fingerprint-based redundancy identifi- cation difficult. Third, very similar but different digital 1 Introduction contents (e.g., files or data streams), which would other- wise present excellent deduplication opportunities, will Due to the information explosion [1, 3], data reduc- become fundamentally distinct compressed packages af- tion technologies such as compression and deduplica- ter applying even the same compression algorithm. -

Software Requirements Specification
Software Requirements Specification for PeaZip Requirements for version 2.7.1 Prepared by Liles Athanasios-Alexandros Software Engineering , AUTH 12/19/2009 Software Requirements Specification for PeaZip Page ii Table of Contents Table of Contents.......................................................................................................... ii 1. Introduction.............................................................................................................. 1 1.1 Purpose ........................................................................................................................1 1.2 Document Conventions.................................................................................................1 1.3 Intended Audience and Reading Suggestions...............................................................1 1.4 Project Scope ...............................................................................................................1 1.5 References ...................................................................................................................2 2. Overall Description .................................................................................................. 3 2.1 Product Perspective......................................................................................................3 2.2 Product Features ..........................................................................................................4 2.3 User Classes and Characteristics .................................................................................5 -

HOW to DOWNLOAD a Gvsig LIVE-DVD from MEGAUPLOAD Some Gvsig Live-Dvds Have Been Uploaded to Megaupload in Several 100MB File
HOW TO DOWNLOAD A gvSIG LIVE-DVD FROM MEGAUPLOAD Some gvSIG Live-DVDs have been uploaded to Megaupload in several 100MB file size. To download all the files at the same time, and mount the DVD image, you must follow the next steps: – Download the free application called JDownloader, that allows to download all the files at the same time. The application can be downloaded from http://jdownloader.org/download/index – Install JDownloader. – Open JDownloader, and go to Link-Add links-Add URL(s) – Click on the LinkGrabber tab, and then click the Continue with all button. The files will be downloaded at the folder configured for it (by default it will be “downloads” at the JDownloader install folder). – Extract the .iso image: – On Windows: – If WinRAR is installed on the computer, it probably recognize the file downloaded, and there will be only a compressed file. Click on it with the right button, and select Extract here. – If the 100MB size files have been downloaded separately, you must download the free application called 7-zip. It can be downloaded from http://www.7- zip.org/. – On Linux: – JDownloader will wownload the 100MB size files separately, and it also will create the .7z compressed file. If there is any compressor installed on the system, to get the .iso image, you only must click on the .7z file with the right button, and then select Extract->Extract here. If there isn't anyone, you can install the free application called 7-zip. It can be installed directly from the package manager, or downloading it from http://www.7-zip.org/. -

Patient Attachment Verification
PATIENT ATTACHMENT VERIFICATION The personal information in this form is collected by the Clinic (named below) and the health authority in which it resides and may be disclosed to the Province’s Ministry of Health under the authority of the Medicare Protection Act and the Freedom of Information and Protection of Privacy Act in order to audit clinic attachments. SECTION A: PERSONAL INFORMATION First Name Last Name Personal Health Number (PHN) SECTION B: CLINIC INFORMATION Practitioner Name Clinic Name SECTION C: PRACTITIONER - PATIENT TERMS Is the Practitioner listed above your Primary Care Provider? Yes No Has the Practitioner listed above had the following conversation outlining the Patient-Practitioner relationship? Yes No Physician: Patient: As your primary care provider I, along with my practice team, As my patient I ask that you: agree to: • Seek your health care from me and my team whenever • Provide you with safe and appropriate care possible and, in my absence, through my colleague(s) • Coordinate any specialty care you may need • Name me as your primary care provider if you have to • Offer you timely access to care, to the best of my ability and as visit an emergency facility or another provider reasonably possible in the circumstances • Communicate with me honestly and openly so we can • Maintain an ongoing record of your health best address your health care needs • Keep you updated on any changes to services offered at my clinic • Communicate with you honestly and openly so we can best address your health care needs SECTION D: AUTHORIZATION I hereby confirm my attachment to the Practitioner listed above. -

7 Zip Latest Version Download 7-Zip Download V18.05
7 zip latest version download 7-Zip Download v18.05. Here you can download the latest version of 7-Zip. 7-Zip is a file archiver with a high compression ratio. You can use 7-Zip on any computer, including a computer in a commercial organization. You don't need to register or pay for 7-Zip. 7-Zip is a file archiver with a high compression ratio for ZIP and GZIP formats, which is between 2 to 10% better than its peers, depending on the exact data tested. And 7-Zip boosts its very own 7z archive format that also offers a significantly higher compression ratio than its peers—up to 40% higher! This is mainly because 7-Zip uses LZMA and LZMA2 compression , with strong compression settings and dictionary sizes , to slow but vastly improve density. If a zip tool gains its appeal from its ability to efficiently compress files, then 7-Zip proves it has a little bit o’ magic. After you effortlessly download and launch 7-Zip, you’ll quickly discover its simple and easy to navigate interface. The main toolbar contains 7- Zip’s most used features and there are several menus that allow you to dig deeper within. For example, the Extract button lets you easily browse for or accept the default destination directory for your file, while the View menu contains a Folder History, and the Favorites menu lets you save up to ten fold ers. 7-Zip also integrates with the Windows Explorer menus, displaying archive files as folders and providing a toolbar with drag-and- drop functions. -

MIGRATORY COMPRESSION Coarse-Grained Data Reordering to Improve Compressibility
MIGRATORY COMPRESSION Coarse-grained Data Reordering to Improve Compressibility Xing Lin*, Guanlin Lu, Fred Douglis, Philip Shilane, Grant Wallace *University of Utah EMC Corporation Data Protection and Availability Division © Copyright 2014 EMC Corporation. All rights reserved. 1 Overview Compression: finds redundancy among strings within a certain distance (window size) – Problem: window sizes are small, and similarity across a larger distance will not be identified Migratory compression (MC): coarse-grained reorganization to group similar blocks to improve compressibility – A generic pre-processing stage for standard compressors – In many cases, improve both compressibility and throughput – Effective for improving compression for archival storage © Copyright 2014 EMC Corporation. All rights reserved. 2 Background • Compression Factor (CF) = ratio of original / compressed • Throughput = original size / (de)compression time Compressor MAX Window size Techniques gzip 64 KB LZ; Huffman coding bzip2 900 KB Run-length encoding; Burrows-Wheeler Transform; Huffman coding 7z 1 GB LZ; Markov chain-based range coder © Copyright 2014 EMC Corporation. All rights reserved. 3 Background • Compression Factor (CF) = ratio of original / compressed • Throughput = original size / (de)compression time Compressor MAX Window size Techniques gzip 64 KB LZ; Huffman coding bzip2 900 KB Run-length encoding; Burrows-Wheeler Transform; Huffman coding 7z 1 GB LZ; Markov chain-based range coder © Copyright 2014 EMC Corporation. All rights reserved. 4 Background • Compression Factor (CF) = ratio of original / compressed • Throughput = original size / (de)compression time Compressor MAX Window size Techniques Example Throughput vs. CF gzip 64 KB LZ; Huffman coding 14 gzip bzip2 900 KB Run-length encoding; Burrows- 12 Wheeler Transform; Huffman coding 10 (MB/s) 7z 1 GB LZ; Markov chain-based range 8 coder Tput 6 bzip2 4 7z 2 0 Compression 2.0 2.5 3.0 3.5 4.0 4.5 5.0 Compression Factor (X) The larger the window, the better the compression but slower © Copyright 2014 EMC Corporation. -

Using LZMA Compression for Spectrum Sensing with SDR Samples
Using LZMA Compression for Spectrum Sensing with SDR Samples Andre´ R. Rosete Kenneth R. Baker Yao Ma Interdisciplinary Telecom. Program Interdisciplinary Telecom. Program Communications Technology Laboratory University of Colorado Boulder University of Colorado Boulder National Institute of Standards and Technology Boulder, Colorado, USA Boulder, Colorado, USA Boulder, Colorado, USA [email protected] [email protected] [email protected] Abstract—Successful spectrum management requires reliable [2], we can expect that communication signal samples will methods for determining whether communication signals are be more compressible than Gaussian noise samples, as the present in a spectrum portion of interest. The widely-used latter are random values. The Lempel-Ziv-Markov chain Algo- energy detection (ED), or radiometry method is only useful in determining whether a portion of radio-frequency (RF) spectrum rithm (LZMA) is a lossless, general-purpose data compression contains energy, but not whether this energy carries structured algorithm designed to achieve high compression ratios with signals such as communications. In this paper we introduce low compute time. [3] We have found that, all else held the Lempel-Ziv Markov-chain Sum Algorithm (LZMSA), a constant, SDR-collected, LZMA-compressed files containing spectrum sensing algorithm (SSA) that can detect the presence in-phase and quadrature (IQ) samples signals with higher of a structured signal by leveraging the Liv-Zempel-Markov chain algorithm (LZMA). LZMA is a lossless, general-purpose signal-to-noise ratios (SNRs) show better compression ratios data compression algorithm that is widely available on many than files containing IQ samples of lower-SNR channels, or of computing platforms. -
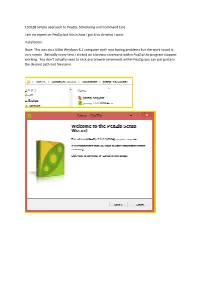
150528 Simple Approach to Peazip, Scheduling and Command Line I
150528 Simple approach to PeaZip, Scheduling and Command Line I am no expert on PeaZip but this is how I got it to do what I want. Installation: Note: This was on a 64bit Windows 8.1 computer and I was having problems but the work round is very simple. Basically every time I clicked on a browse command within PeaZip the program stopped working. You don’t actually need to click any browse commands within PeaZip you can just paste in the desired path and file name. No System integration: By default PeaZip incorporates itself into your file explorer, when you right mouse click on any file the PeaZip option is listed. If you need this OK otherwise NO. Next> will confirm your selections and allow you to continue the installation. Creating an Archive: Desktop> Highlight the directory to be Archived and Right Mouse click> Add> To create unique Archives each time click the Append Timestamp to Archive. Click OK and PeaZip will create your backup. Above is a one off Archive, what if you want to schedule the archive to run each day. Change the Task Name to something that makes sense to you. Set your start time. Add Schedule> YES do this now but you will have to edit the Archive_Codecademy04_test02.bat later to get the date time stamp to work properly. C:\Users\Controller\AppData\Roaming\PeaZip\Scheduled scripts That is it – your PeaZip has been scheduled to run at 12:00 every working day. There is one problem – It will have the same name every time, the date time stamp is as was originally created.