Department of Computational Biology and Medical Sciences Department
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
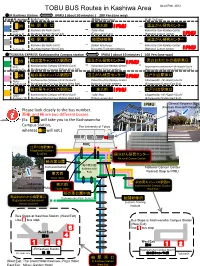
TOBU BUS Routes in Kashiwa Area As of Feb. 2012
TOBU BUS Routes in Kashiwa Area As of Feb. 2012 ◎JR Kashiwa Station IPMU ( about 30 minutes ) 280 Yen (one way) 系統番号 Number 起点 From / To 経由地 Other Stops 起点 To/From 西 01 柏 駅 西 口 東大前 IPMU ― 国立がん研究センター 柏 ― Nishi- Kashiwa-Eki Nishi-Guchi Todai Mae Kokuritsu-Gan-Kenkyu-Center Kashiwa 01 (Kashiwa Station West Exit) (Tokyo university) (National Cancer center) IPMU 柏 駅 西 口 税関研修所 国立がん研究センター 柏 44 ― Kashiwa-Eki Nishi-Guchi ― Zeikan Kenshu-jo Kokuritsu-Gan-Kenkyu-Center Kashiwa 44 (Kashiwa Station West Exit) (Customs Training Institute) (National Cancer center) IPMU ◎TSUKUBA EXPRESS Kashiwanoha Campus station IPMU ( about 10 minutes ) 160 Yen (one way) 西 柏の葉キャンパス駅西口 国立がん研究センター 流山おおたかの森駅東口 03 柏 IPMU ― ― Nishi- Kashiwanoha Campus-Eki Nishi-Guchi Kokuritsu-Gan-Kenkyu-Center Nagareyama-otakanomori-Eki Higashi-Guchi Kashiwa 03 (Kashiwanoha Campus Station West Exit) (National Cancer center) (Nagareyama-otakanomori Station East Exit) 西 柏の葉キャンパス駅西口 国立がん研究センター 江戸川台駅東口 04 柏 IPMU Nishi- Kashiwanoha Campus-Eki Nishi-Guchi Kokuritsu-Gan-Kenkyu-Center Edogawadai -Eki Higashi-Guchi Kashiwa 04 (Kashiwanoha Campus Station West Exit) (National Cancer center) (Edogawadai Station East Exit) 西 柏の葉キャンパス駅西口 東大前 IPMU 江戸川台駅東口 10 柏 ― ― Kashiwanoha Campus-Eki Nishi-Guchi Todai Mae Edogawadai -Eki Higashi-Guchi Nishi- Kashiwa 10 (Kashiwanoha Campus Station West Exit) (Tokyo university) (Edogawadai Station East Exit) IPMU General Research Bldg. (Kashiwa Research Complex) Please look closely to the bus number. 西柏 and 柏 are two different busses. 西 (Ex. 02 will take you to the -

東京大学大気海洋研究所 Atmosphere and Ocean Research Institute, the University of Tokyo / ANNUAL REPORT
東京大学海洋研究所要覧2020 /表紙 ATMOSPHERE AND OCEAN RESEARCH INSTITUTE, THE UNIVERSITY OF TOKY O | 2020 CATALO 東京大学大気海洋研究所 G Atmosphere and Ocean Research Institute, The University of Tokyo / ANNUAL REPORT 要覧|CATALOG 2020 年報|ANNUAL REPORT CONTENTS 2020 ATMOSPHERE AND OCEAN RESEARCH INSTITUTE THE UNIVERSITY OF TOKYO 要覧|CATALOG P2 年報|ANNUAL REPORT P74 沿革 P 国際協力 P History 2 International Cooperation 75 機構 P 共同利用研究活動 P Organization 4 Cooperative Research Activities 87 委員会 P 教育活動 P Committees 6 Educational Activities 101 教職員 P 予算 P Staff 8 Budget 104 共同利用・共同研究拠点 P 研究業績 P Joint Usage / Research Center 13 Publication List 105 教 育システム P Educational System 23 部門とセンターの研究内容 P Research Contents 28 COVER IMAGES ❶:プレスリリース「モガニ属の新種『オオヨツハモ ❻:研究トピックス「単細胞を飼う単細胞:浮遊性有 ガニ』を発見~三陸の藻場における重要種」か 孔虫と藻類の光共生関係の解明」から「本研究 ら「 新 種 オ オ ヨ ツ ハ モ ガ ニ P. ferox Ohtsuchi & で用いた浮遊 性有孔虫3 0 種。スケールバーは Kawamura, 2019」 20 0 μm」 1 2 ❷:プレスリリース「ウミガメ由来の海洋観測データ ❼:UTokyo FOCUS – Articles「海海洋の内部波に 3 を季節予測シミュレーションに活用 ―バイオロ よるサンゴ礁の冷却 ~白化緩和効果の可能性 ギング手法により海洋・気象観測網の発展に可 を指摘~」から”Researchers maintain a reef 4 5 能性―」から「産卵を終えた後、深度・水温情報 flat temperature logger in Iriomote, Japan. を送信する人工衛星対応型発信器を背甲に取 Photo by Alex S.J. Wyatt.” り付けられて海に帰るヒメウミガメ。2019年6月、 ❽:研究トピックス「オオメジロザメはなぜ河川でも生 インドネシア 西 パ プ ア 州 のワルマメディ海 岸に 息できるのか? その生理学的メカニズムの解明」 6 7 8 て。装置はエポキシ接着剤で装着され、1年から2 から「研究対象のオオメジロザメ。沖縄本島や西 年後に自然に脱落する。現地政府の許可を受け 表 島などの 河 川でも生 息が 確 認されている」 て野外調査を行っている。(撮影:佐藤克文)」 ❾:2019年に就航30周年を迎えた学術研究船「白 ❸:AORI写真コンテスト2019海・空部門エントリー 9 1 0 11 鳳 丸 」( 2 代 目 ) 作品「富士山・笠雲・レンズ雲」(一部抜粋)松峯 正典 1 0:研究トピックス「海底堆積物中の有孔虫化石の 高精度分析から読み解く大気・海洋間の炭素交 ❹:研 究トピックス「インドネシアシーラカンス研 究 最 換 ~ 最 終氷期からの回 復 期に赤 道 太平洋から 前 線 」から「インドネシアシーラカンス計 測の様 大気への二酸化炭素放出は不均質に起きていた 子。2017年5月31日 アクアマリンふくしまにて」 ことを解明~」から「本研究の分析に用いた浮遊 ❺:HPCI戦略プログラム分野3の一環としてスー 性有孔虫G. -

Second International SALSA Workshop
Second International SALSA Workshop Development of Seamless chemical AssimiLation System and its Application for atmospheric environmental materials (Project SALSA) Invited Speaker Prof. James Schauer: Department of Civil and Environmental Engineering University of Wisconsin Prof. Soon-Chang Yoon: School of Earth and Environmental Sciences Seoul National University Prof. Xiquan Dong: Department of Atmospheric Sciences University of North Dakota Dr. Konstantin Gribanov: Department of Physics Ural Federal University 15-16 December 2011 Room 257 (Auditorium) Atmosphere and Ocean Research Institute (AORI) The University of Tokyo Kashiwanoha, Kashiwa JAPAN Access http://www.aori.u-tokyo.ac.jp/access/index.html Program Dec. 15 Date Time Schedule 10:00 - 10:05 Opening (Chair: T. Inoue(AORI)) 10:05 -10:10 Welcome: T. Nakajima (AORI) Group 0: Managing 10:10 - 10:40 G0_1: T. Nakajima (AORI) 10:40 - 11:10 Invited: S.-C. Yoon (SNU: Korea) 11:10 - 11:40 Invited: X. Dong (UND: USA) 11:40 - 13:20 Lunch Group 2: Air pollutant assimilation 13:20 - 13:40 G2_1: M. Uematsu (AORI) (Chair:H. Tsuruta (AORI)) 13:40 - 14:00 G2_2: J. Uchida (AORI) 14:00 - 14:20 G2_3: T. Dai (AORI from China) 12/15 14:20 - 14:40 G2_4: D. Goto (AORI) (Thu) 14:40 - 15:00 G2_5: T. Ohara (NIES) (talked by H. Tsuruta (AORI)) 15:00 - 15:10 Break Group 3: Impact assessment group 15:10 - 15:40 Invited: J. Schauer (UW: USA) 15:40 - 16:00 G3_1: A. Takami (NIES) (Chair) 16:00 - 16:20 G3_2: K. Ueda (NIES) 16:20 - 16:30 Break 16:30 - 16:50 G3_3: A. -

Welcome to Kashiwa Campus the University of Tokyo
Welcome to Kashiwa Campus The University of Tokyo Adventures in Knowledge Kashiwa Campus is the newest of the three main campuses of The University of Tokyo in addition to campuses at Hongo and Komaba, and was established with the vision of building a new community with a strong interdisciplinary and international character that can respond to the dynamic scientific and educational needs of the 21st Century. Hongo Campus, the core of traditional academic activities at The University of Tokyo since its foundation in 1877, and Komaba Campus, focusing on undergraduate studies and building interdisciplinary links between established fields of study for over 60 years, have longstanding academic traditions. Kashiwa Campus recently joined this family and aims at a fundamental reorganization of the structure of traditional academic disciplines, reconsidering the framework of how knowledge is created and organized. Under the slogan of “Adventures in Knowledge”, Kashiwa Campus has started various new initiatives. The Graduate School of Frontier Sciences (GSFS) consists of three divisions each with relevant departments: Division of Transdisciplinary Sciences, Division of Biosciences, and Division of Environmental Studies. GSFS aims to create new frontiers of academic fields. The Institute for Solid State Physics (ISSP) is exploring a wide variety of phenomena exhibited by various materials from the viewpoint of basic science using state-of-art experimental facilities and techniques. Materials under study include those forming the basis of today's high-tech society and its future development. The Institute for Cosmic Ray Research (ICRR) is pursuing the origins of the universe and matter through research on cosmic rays. The Atmosphere and Ocean Research Institute (AORI) not only promotes basic research on the ocean and atmosphere, but also contributes to society through deepening ocean and climate science. -

Welcome to Kashiwa Campus the University of Tokyo
Welcome to Kashiwa Campus The University of Tokyo Adventures in Knowledge Kashiwa Campus is the newest of the three main campuses of The University of Tokyo in addition to campuses at Hongo and Komaba, and was established with the vision of building a new community with a strong interdisciplinary and international character that can respond to the dynamic scientific and educational needs of the 21st Century. Hongo Campus, the core of traditional academic activities at The University of Tokyo since its foundation in 1877, and Komaba Campus, focusing on undergraduate studies and building interdisciplinary links between established fields of study for over 60 years, have longstanding academic traditions. Kashiwa Campus recently joined this family and aims at a fundamental reorganization of the structure of traditional academic disciplines, reconsidering the framework of how knowledge is created and organized. Under the slogan of “Adventures in Knowledge”, Kashiwa Campus has started various new initiatives. The Graduate School of Frontier Sciences (GSFS) consists of three divisions each with relevant departments: Division of Transdisciplinary Sciences, Division of Biosciences, and Division of Environmental Studies. GSFS aims to create new frontiers of academic fields. The Institute for Solid State Physics (ISSP) is exploring a wide variety of phenomena exhibited by various materials from the viewpoint of basic science using state-of-art experimental facilities and techniques. Materials under study include those forming the basis of today's high-tech society and its future development. The Institute for Cosmic Ray Research (ICRR) is pursuing the origins of the universe and matter through research on cosmic rays. The Atmosphere and Ocean Research Institute (AORI) not only promotes basic research on the ocean and atmosphere, but also contributes to society through deepening ocean and climate science. -

IPMU Brochure
Institute for the Physics and Mathematics of the Universe Todai Institutes for Advanced Study The University of Tokyo The universe is written in the language of mathematics These words of Galileo, who lived at the dawn of cosmology in the 16 to 17th century, give an important foresight to the strategy we must take at IPMU to answer big questions we face today. IPMU addresses deep mysteries of the universe by integrating the forefront knowledge of physics and mathematics. New approaches under new strategies open new possibilities. World Premier International Research Center Initiative Our universe is full of mystery. “Everything is made of atoms” had been the basic concept that had dominated our view of the universe for the past two centuries. The universe was made of atoms that obeyed quantum mechanics, and all gravity phenomena on earth and in the solar system were well-described by Einstein’s general relativity. But the 1998 discovery that “the universe is expanding with an accelerated speed” has changed our entire understanding. Many researchers now believe that mysterious dark energy must be distributed across the universe and the cause of this acceleration. Atoms, human, solar system The universe is expanding with an accelerated speed. ©NASA, ESA, J. Blakeslee and H. Ford (JHU) It has been known for some time that the galaxies must contain large amount of invisible dark matter in order to hold their spiral shapes. But we do not know what they are. We knew only 5% of the universe! Atoms: 4.4% Stars: 0.5% Neutrinos: 0.1%? Dark Matter: 22% Does not emit light and invisible to us, but needed for holding the galaxies together. -
Welcome to Kashiwa Campus the University of Tokyo
Welcome to Kashiwa Campus The University of Tokyo Adventures in Knowledge Kashiwa Campus is the newest of the three main campuses of The University of Tokyo in addition to campuses at Hongo and Komaba, and was established with the vision of building a new community with a strong interdisciplinary and international character that can respond to the dynamic scientific and educational needs of the 21st Century. Hongo Campus, the core of traditional academic activities at The University of Tokyo since its foundation in 1877, and Komaba Campus, focusing on undergraduate studies and building interdisciplinary links between established fields of study for over 60 years, have longstanding academic traditions. Kashiwa Campus recently joined this family and aims at a fundamental reorganization of the structure of traditional academic disciplines, reconsidering the framework of how knowledge is created and organized. Under the slogan of “Adventures in Knowledge”, Kashiwa Campus has started various new initiatives. The Graduate School of Frontier Sciences (GSFS) consists of three divisions each with relevant departments: Division of Transdisciplinary Sciences, Division of Biosciences, and Division of Environmental Studies. GSFS aims to create new frontiers of academic fields. The Institute for Solid State Physics (ISSP) is exploring a wide variety of phenomena exhibited by various materials from the viewpoint of basic science using state-of-art experimental facilities and techniques. Materials under study include those forming the basis of today's high-tech society and its future development. The Institute for Cosmic Ray Research (ICRR) is pursuing the origins of the universe and matter through research on cosmic rays. The Atmosphere and Ocean Research Institute (AORI) not only promotes basic research on the ocean and atmosphere, but also contributes to society through deepening ocean and climate science. -

AIST-Utokyo Advanced Operando-Measurement Technology Research and Open Innovation Laboratory Research and (OPERANDO-OIL) Testing Complexⅱ Testing Complexⅰ
Access Map of Kashiwa Campus, The University of Tokyo N AIST-UTokyo Advanced Operando-Measurement Technology Research and Open Innovation Laboratory Research and (OPERANDO-OIL) Testing ComplexⅡ Testing ComplexⅠ CChibahiba EExperimentxperiment SStation,tation, IIISIS Laser and Synchrotron Research Laboratory, ISSP National Institute of Advanced Industrial Science and Technology (AIST) Ocean Observation Transdisciplinary Science Department of Materials and Chemistry Warehouse, AORI Laboratory, GSFS Low-Rise Building, ISSP Kashiwa Research Complex 2 Institute for Cosmic Todai Kashiwa Venture Plaza Environmental Institute for Solid State Ray Research (ICRR) Biosciences, Transdisciplinary Studies, GSFS Physics (ISSP) Kavli IPMU Tokatsu Techno Plaza GSFS Science, GSFS Kashiwa Research Atmosphere and Ocean Complex Research Institute (AORI) Restaurant Kashiwa Health (Plaza Ikoi ) Service Center Cafeteria Kashiwa Guest House Academic Sushi-Restaurant (Hama) Shop Food shop & Cafe Kashiwa Library Gatehouse Gorokurō Pond Main Gate AIST-UTokyo By Train Advanced Operando-Measurement Technology To Tsukuba - From Kashiwanoha-campus station (west entrance) of TSUKUBA To Kasukabe EXPRESS (TX) Line Open Innovation Laboratory Joban High Way Tobu Urban Park L. Kashiwa IC ・Tobu Bus “Nishikashiwa03 (西柏03)” for east entrance of Nagareya- Edogawadai Sta. ma-otakanomori Station → get off at “National Cancer Center” Route 16 stop. ・Tobu Bus “Nishikashiwa04 (西柏04)” for east entrance of Edogawa- The University of Tokyo dai Station → get off at “National Cancer Center” stop. To Tokyo Kashiwa Campus ・Tobu Bus “Nishikashiwa10 (西柏10)” for east entrance of Edogawa- dai Station → get off at “Todai-mae” stop. The University of Tokyo ・About 25 minutes' walk from Kashiwanoha-campus station to the campus Kashiwanoha Campus - From Kashiwa station (west entrance) of JR Joban Line Station Satellite (Future Center) ・Tobu Bus “Nishikashiwa01 (西柏01)” or “Kashiwa44(柏44)” for sukuba National Cancer Center → get off at “National Cancer Center” stop. -

Kashiwa Campus Tsukuba Express Kashiwanoha Campus Sta.(West Exit) (Walk 3Min
2016.02 Using the Joban Expressway, get off at the Kashiwa Kashiwa I.C. I.C. for Chiba direction (go right after the toll gate) on to the rout 16. Change to the right lane soon. Drive 500 m. and turn right at the first traffic light. Kashiwa Campus Tsukuba Express Kashiwanoha Campus Sta.(West exit) (Walk 3min. ) Nishi-kashiwa 03 Nagareyama-Otakanomori-eki higashi guchi Todai mae or Todai nisi Tobu Bus Todai nishi 10min. Nishi-kashiwa 04/Nishi-kashiwa 10 National Edogawadai-eki higashi guchi Cancer Center SHUTTLE BUS (UT-Kashiwa Campus - Kashiwanoha Campus Station) 6min. Taxi 5min. Walk 25min. Tobu Noda Line Tsukuba Express Kashiwanoha Highway bus from Narita Airport 75-100 min. Campus JR Musashino Line Kashiwanoha Campus Sta. 2min. Tobu Highway bus 75min. Nagareyama- otakanomori JR Joban-Line / Tobu Noda-Line Kashiwa Sta. (West exit) 5min. 5min. Yaman Nishi-kashiwa 01 JR ote L ine Minami- bound for National Cancer Center via Kashiwa Park 25min. nagareyama Tobu Bus Taxi 20min. 8HQRĺ.DVKLZD (JR Joban Line rapid service) 29 min. Ueno Kashiwa JR Joban Line 9min. 17min. $NLKDEDUDĺ.DVKLZDQRKD&DPSXV Akihabara 33min. Higashimatsudo 9min. Shinkamagaya 1DULWD$LUSRUWĺ6KLQNDPDJD\DPLQ Hamamatsucho +DPDPDWVXFKRĺ$NLKDEDUDPLQ 1DULWD$LUSRUWĺ+LJDVKLPDWVXGRPLQ +DPDPDWVXFKRĺ8HQRPLQ Keisei Sky Access Express 1LVKLIXQDEDVKLĺ )XQDEDVKLĺ.DVKLZDPLQ Minami-nagareyama 18min. )XQDEDVKLĺ1DJDUH\DPDRWDNDQRPRULPLQ Tokyo Monorail 30 min. Nishi-funabashi JR Sobu Line 3 min. Funabashi Keisei Honsen Line limited express 49 min. Narita Airport Haneda Airport Administrative Office, Graduate School of Frontier Sciences 5-1-5 Kashiwanoha, Kashiwa-shi, Chiba-ken 277-8561 Tel. 04 -7136- 4003 Fax 04 -7136-4020 Graduate School of Frontier Sciences, The University of Tokyo. -

Center for Foreign Residents Shown Hereunder. the Support Center For
Welcome to Nagareyama City! Dear Citizens of Foreign Nationals, We, NIFA’s members of Foreign Residents Support Group, have been trying to support and assist you in your everyday life in Nagareyama, mainly by providing you with the information to be required for your safe and pleasant life here. So far, we have been posting some information on Emergencies (Fire, Police and Earthquake), Evacuations Sites, Hospitals / Clinics, Household Garbage Sorting–out /Disposal and such on our Home Page for quite some time now. This time, we have uploaded revised information according to the latest edition of “Shimin Benricho ” (Citizen’s Handbook) updated by Nagareyama City Office on their Website on June 1, 2019. Please note this English version is a combination of English translations of the original Japanese texts and some additional extended information for your better understandings. There will be others to follow, if necessary. So please give us your requests and comments for/on what information you need further for your daily life in Nagareyama by phone or E-mail to The Support Center for Foreign Residents shown hereunder. We sincerely hope the information shown in this volume will be of some service to you all. Best regards, Foreign Residents Support Group, Nagareyama International Friendship Association (NIFA) The Support Center for Foreign Residents: Telephone: 04-7128-6007 E-mail address: [email protected] 1 Nagreyama Citizen’s Handbook CONTENTS: 1. The Access to the Nagareyama City Office (Head Office) P3 and The Services of Each Section 2. The Access to the Branch Offices and Services P4 3. -

Explanatory Leaflet Department of Advanced Energy
YEAR 2019 Master course Doctor course Explanatory Leaflet Graduate School of Frontier Sciences Department of Advanced Energy Nuclear Fusion Research Education Program Examination date and time Master course August 21 (Tue),August 22 (Wed),August 28 (Tue), 2018 Doctor course August 21 (Tue),August 22 (Wed),August 23 (Thu), 2018 Guidance May 25 (Fri) 16:30~18:00, 2018 (Hongo Campus) Eng. Bldg. No.2, 10F Meeting Room 5 May 26 (Sat) 13:00~15:00, 2018 (Kashiwa Campus) Kashiwa Library, Media Hall Contact Kashiwanoha 5-1-5, Kashiwa-shi, Chiba 277-8561 Kiban Bldg. 1F The University of Tokyo Graduate school of Frontier Sciences, Kyomu-kakari TEL 04-7136-4092 OR Contact Person of Department of Advanced Energy e-mail : [email protected] Homepage http://www.k.u-tokyo.ac.jp/ae/ * This document is a translation of necessary part of the Japanese version. In the event that any question should arise about this version, the Japanese version is the authoritative version INDEX Department of Advanced Energy ........................................... 2 [1]Preface .............................................................................................................................................. 2 [2]Research fields ................................................................................................................................. 2 [3]Special Selection for Master course ............................................................................................... 9 [4]Special Selection for Doctor course .............................................................................................