Sorting out the Genetic Background of the Last Surviving South China
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Rev Cop16) on Conservation of and Trade in Tiger
SC65 Doc. 38 Annex 1 REVIEW OF IMPLEMENTATION OF RESOLUTION CONF. 12.5 (REV. COP16) ON CONSERVATION OF AND TRADE IN TIGERS AND OTHER APPENDIX-I ASIAN BIG CAT SPECIES* Report to the CITES Secretariat for the 65th meeting of the Standing Committee Kristin Nowell, CAT and IUCN Cat Specialist Group and Natalia Pervushina, TRAFFIC With additional support from WWF Table of Contents Section Page Executive Summary 2 1. Introduction 8 2. Methods 11 3. Conservation status of Asian big cats 12 4. Seizure records for Asian big cats 13 4.1. Seizures – government reports 13 4.1.1. International trade databases 13 4.1.2. Party reports to CITES 18 4.1.3. Interpol Operation Prey 19 4.2. Seizures – range State and NGO databases 20 4.2.1. Tigers 21 4.2.2. Snow leopards 25 4.2.3. Leopards 26 4.2.4. Other species 27 5. Implementation of CITES recommendations: best practices and 28 continuing challenges 5.1. Legislative and regulatory measures 28 5.2. National law enforcement 37 5.3. International cooperation for enforcement and conservation 44 5.4. Recording, availability and analysis of information 46 5.5. Demand reduction, education, and awareness 48 5.6. Prevention of illegal trade in parts and derivatives from captive 53 facilities 5.7. Management of government and privately-held stocks of parts and 56 derivatives 5.8. Recent meetings relevant to Asian big cat conservation and trade 60 control 6. Recommendations 62 References 65 * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. -

Opportunity for Thailand's Forgotten Tigers: Assessment of the Indochinese Tiger Panthera Tigris Corbetti and Its Prey with Camera-Trap Surveys
Opportunity for Thailand's forgotten tigers: assessment of the Indochinese tiger Panthera tigris corbetti and its prey with camera-trap surveys E RIC A SH, Ż ANETA K ASZTA,ADISORN N OOCHDUMRONG,TIM R EDFORD P RAWATSART C HANTEAP,CHRISTOPHER H ALLAM,BOONCHERD J AROENSUK S OMSUAN R AKSAT,KANCHIT S RINOPPAWAN and D AVID W. MACDONALD Abstract Dramatic population declines threaten the En- Keywords Bos gaurus, distribution, Dong Phayayen-Khao dangered Indochinese tiger Panthera tigris corbetti with ex- Yai Forest Complex, Indochinese tiger, Panthera tigris tinction. Thailand now plays a critical role in its conservation, corbetti, prey abundance, Rusa unicolor, Sus scrofa as there are few known breeding populations in other Supplementary material for this article is available at range countries. Thailand’s Dong Phayayen-Khao Yai For- doi.org/./S est Complex is recognized as an important tiger recovery site, but it remains poorly studied. Here, we present results from the first camera-trap study focused on tigers and im- plemented across all protected areas in this landscape. Our Introduction goal was to assess tiger and prey populations across the five protected areas of this forest complex, reviewing discernible he tiger Panthera tigris has suffered catastrophic de- patterns in rates of detection. We conducted camera-trap Tclines in its population (%) and habitat (%) over surveys opportunistically during –. We recorded the past century (Nowell & Jackson, ; Goodrich et al., , detections of tigers in , camera-trap nights. ; Wolf & Ripple, ). Evidence suggests only source Among these were at least adults and six cubs/juveniles sites (i.e. sites with breeding populations that have the po- from four breeding females. -

The Oriental Pearl Radio & TV Tower 东方明珠 Getting in Redeem Your
The Oriental Pearl Radio & TV Tower 东方明珠 Getting In Redeem your pass for an admission ticket at the first ticket office, near No. 1 Gate. Hours Daily, 8:00 am-9:30 pm. Address No. 1 Lujiazui Century Ave Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, get out from Exit 1 and walk to The Oriental Pearl Radio & TV Tower. Yu Garden (Yuyuan) 豫园 Getting In Please redeem your pass for an admission ticket at the Yuyuan Garden ticket office located on the north side of the Huxin Pavilion Jiuqu Bridge prior to entry. Hours Daily, 8:45 am-4:45 pm. Address No. 218 Anren St Huangpu District, Shanghai Public Transportation Take Metro Line 10 and get off at Yuyuan Station, then walk to Yu Garden. Shanghai World Financial Center Observatory 上海环球金融中心 Getting In Please redeem your pass for an admission ticket at the Global Finance Center F1 ticket window located at Lujiazui Century Ave. Hours Daily, 9:00 am-10:30 pm. Address B1 Ticketing Window, World Financial Center 100 Century Avenue Lujiazui, Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, then walk to Shanghai World Financial Center. Shanghai Hop-On Hop-Off Sightseeing Bus Tour 观光巴士 Getting In You must first redeem your pass for a bus ticket at one of the following locations prior to boarding: Nanjing Road Station (New World City Stop): Opposite to New World City, No. 2-88 Nanjing West Road, Huangpu District, Shanghai Bund A Station (Sanyang Food Stop): Beside Sanyang Food, 367 East Zhongshan Road, Huangpu District, Shanghai (near Beijing East Road) Shiliupu Station (Pujiang Tour Terminal Stop): 531 Zhongshan East Second Road, Huangpu District, Shanghai Yuyuan Station (Yongan Road, Renmin Road): Xinkaihe Road, Renmin Road, next to the bus stop in front of the Bund soho. -

Tigers Lǎohǔ 老 虎
◀ Tibetans (Zang) Comprehensive index starts in volume 5, page 2667. Tigers Lǎohǔ 老 虎 Once a royal symbol of war, tigers have be- the emperor and empress. The Asian equivalent of the come a casualty of both traditional Chinese lion as the “king of the jungle,” the strong and elegant medicine and environmental destruction in tiger has also been an important icon as the White Tiger modern China, fading into extinction. The of the West, one of the Four Constellations of Chinese astronomy, and a prevalent image in Buddhist lore and most endangered species of tiger, the South martial arts such as Shaolin. China Tiger, has not been spotted in the wild Although the Indochinese tiger (Panthera tigris cor- since the 1960s, and only a few dozen survive in betti) is found in China, Cambodia, Laos, Myanmar captivity, making it “functionally extinct.” (Burma), Thailand, and Vietnam (the International Union for Conservation of Nature and Natural Resources esti- mates that only 630 survive), the South China or Amoy eeply ingrained in Chinese culture as a fierce Tiger (Panthera tigris amoyensis), is the indigenous spe- symbol of war, tigers were, for millennia, the cies with which most Chinese relate. With no official emblems of the highest ministers of defense sighting since 1964, the Amoy has faded into “functional in China, second only to the dragon and the phoenix of extinction” since the 1950s, when at least four thousand A paper cut-out depicting Wu Sung, the folk hero from the famous Chinese novel Water Margin, who was revered for killing the tiger that had terrified the people living on Mount Ching Yang. -

Bibliography on Tiger (Panthera Tigris L.)
Bibliography on Tiger (Panthera tigris L.) Global Tiger Forum Publication 2014 Copyright © Secretariat of Global Tiger Forum, 2014 Suggested Citation: Gopal R., Majumder A. and Yadav S.P. (Eds) (2014). Bibliography on Tiger (Panther tigris L.). Compiled and published by Global Tiger Forum, p 95. Cover Pic Vinit Arora Inside pictures taken by Vinit Arora, Samir K. Sinha, Aniruddha Majumder and S.P.Yadav CONTENTS Acknowledgements i Introduction to Bibliography on tiger 1 Literature collection and compilation process for bibliography on tiger 2-4 1) Ecology, Natural History and Taxonomy 5-23 2) Aspects of Conflicts 24-35 3) Monitoring (tiger, co-predator, prey and habitat) and Status 36-62 evaluation 4) Genetics, morphology, health and disease monitoring 63-75 5) Protection, Conservation, Policies and Bio-politics 76-95 Acknowledgements The “Bibliography on Tiger (Panthera tigris L.)” is an outcome of the literature database on tiger, brought out by the Global Tiger Forum (GTF). The GTF is thankful to all officials, scientists, conservationists from 13 Tiger Range Countries for their support. Special thanks are due to Dr Adam Barlow, Mr. Qamar Qureshi, Dr. Y.V. Jhala, Dr K. Sankar, Dr. S.P. Goyal, Dr John Seidensticker, Dr. Ullas Karanth, Dr. A.J.T Johnsingh, Dr. Sandeep Sharma, Ms. Grace Gabriel, Dr. Sonam Wangchuk, Mr Peter Puschel, Mr. Hazril Rafhan Abdul Halim, Mr Randeep Singh and Dr. Prajna Paramita Panda for sharing some important references on tiger. Mr P.K. Sen, Dr Jagdish Kiswan, Mr Vivek Menon, Mr Ravi Singh and Dr Sejal Vora and Mr Keshav Varma are duly acknowledged for their comments and suggestions. -

The Oriental Pearl Radio & TV Tower 东方明珠
The Oriental Pearl Radio & TV Tower 东方明珠 Hours: Daily, 9:00 am-9:30 pm. Address: No. 1 Century Ave Pudong New Area (Lujiazui), Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, get out from Exit 1 and walk to The Oriental Pearl Radio & TV Tower. Getting In Redeem your pass for an admission ticket at the first ticket office, near No. 1 Gate: Shanghai World Financial Center Observatory 上海环球金融中心 Hours: Daily, 9:00 am-10:00 pm. Address: B1 Ticketing Window, World Financial Center 100 Century Avenue Lujiazui, Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, then walk to Shanghai World Financial Center. Getting In Please redeem your pass for an admission ticket at B1 Ticketing Window, World Financial Center at Lujiazui Century Ave: Pujiang River Cruise Tour 黄浦江“清游江”游览船 Hours:Daily, 10:00 am-8:30 pm. Address:Shiliupu Cruise Terminal,No. 481 Zongshan Rd,Huangpu District, Shanghai Public Transportation Bus: Take the bus #33, 55, 65, 305, 868, 910, 926 or 928 and get off at the Xinkaihe Road-Bus Stop of Zhongshan East Second Road, then walk to No. 481, Zhongshan East Second Road, Huangpu District. Getting In Redeem your pass for an admission ticket at the Shiliu Pu Pier, Huangpu River Tour ticket window at 481 Zhongshan 2nd Rd: Yu Garden (Yuyuan) 豫园 Hours: Daily, 8:45 am-4:45 pm. Address: No. 218 Anren St Huangpu District, Shanghai Public Transportation Take Metro Line 10 and get off at Yuyuan Station, then walk to Yu Garden. -
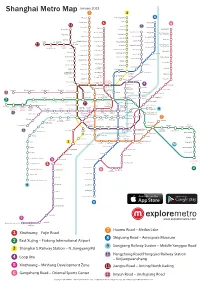
Shanghai Metro Map 7 3
January 2013 Shanghai Metro Map 7 3 Meilan Lake North Jiangyang Rd. 8 Tieli Rd. Luonan Xincun 1 Shiguang Rd. 6 11 Youyi Rd. Panguang Rd. 10 Nenjiang Rd. Fujin Rd. North Jiading Baoyang Rd. Gangcheng Rd. Liuhang Xinjiangwancheng West Youyi Rd. Xiangyin Rd. North Waigaoqiao West Jiading Shuichan Rd. Free Trade Zone Gucun Park East Yingao Rd. Bao’an Highway Huangxing Park Songbin Rd. Baiyin Rd. Hangjin Rd. Shanghai University Sanmen Rd. Anting East Changji Rd. Gongfu Xincun Zhanghuabang Jiading Middle Yanji Rd. Xincheng Jiangwan Stadium South Waigaoqiao 11 Nanchen Rd. Hulan Rd. Songfa Rd. Free Trade Zone Shanghai Shanghai Huangxing Rd. Automobile City Circuit Malu South Changjiang Rd. Wujiaochang Shangda Rd. Tonghe Xincun Zhouhai Rd. Nanxiang West Yingao Rd. Guoquan Rd. Jiangpu Rd. Changzhong Rd. Gongkang Rd. Taopu Xincun Jiangwan Town Wuzhou Avenue Penpu Xincun Tongji University Anshan Xincun Dachang Town Wuwei Rd. Dabaishu Dongjing Rd. Wenshui Rd. Siping Rd. Qilianshan Rd. Xingzhi Rd. Chifeng Rd. Shanghai Quyang Rd. Jufeng Rd. Liziyuan Dahuasan Rd. Circus World North Xizang Rd. Shanghai West Yanchang Rd. Youdian Xincun Railway Station Hongkou Xincun Rd. Football Wulian Rd. North Zhongxing Rd. Stadium Zhenru Zhongshan Rd. Langao Rd. Dongbaoxing Rd. Boxing Rd. Shanghai Linping Rd. Fengqiao Rd. Zhenping Rd. Zhongtan Rd. Railway Stn. Caoyang Rd. Hailun Rd. 4 Jinqiao Rd. Baoshan Rd. Changshou Rd. North Dalian Rd. Sichuan Rd. Hanzhong Rd. Yunshan Rd. Jinyun Rd. West Jinshajiang Rd. Fengzhuang Zhenbei Rd. Jinshajiang Rd. Longde Rd. Qufu Rd. Yangshupu Rd. Tiantong Rd. Deping Rd. 13 Changping Rd. Xinzha Rd. Pudong Beixinjing Jiangsu Rd. West Nanjing Rd. -

Sumatran Tiger
[ABCDE] Volume 1, Issue 7 Nov. 6, 2001 CURRICULUM GUIDE: TIGERS e r I n E d u c a p a p t i o w s n P N e r o t g s r a P o m n t o g i n h s T a h e W C e u h r T r i f c u O l u e r m o C A t e T h h T e t C A o r m KLMNO e u l O u An Integrated Curriculum c f i r Resource Program T r h u e C W e a h s T h i n g t o n P m o a s r t g N o r e P w s n p o a i p t a e c r u I d n E ASSOCIATED PRESS PHOTO IN THIS ISSUE Word Study Tiger Resources Wild Vocabulary 2 4 7 A look at extinction Tigers in Print Academic Content 3 5 Endangered Species 8 Standards © 2001 The Washington Post Company An Integrated Curriculum For The Washington Post Newspaper In Education Program KLMNO Volume 1, Issue 7 Nov. 6, 2001 Sumatran Tiger Tiger Resources KidsPost Article: "Earning His Stripes” On the Web and in Print ON THE WEB http://www.fonz.org/animals/tigertiger/tiger- Lesson: Investigating rare and endangered animals cubpr1.htm Level: Intermediate Friends of the National Zoo Subjects: Science See pictures and learn about Sumatran tigers at the zoo. Related Activity: Geography, English, Language Arts www.5tigers.org Procedure 5 Tigers: The Tiger Information Center Dedicated to providing an international forum focusing Read and Discuss on the preservation of wild tigers across Asia and in Read the KidsPost article. -

De Grave & Fransen. Carideorum Catalogus
De Grave & Fransen. Carideorum catalogus (Crustacea: Decapoda). Zool. Med. Leiden 85 (2011) 407 Fig. 48. Synalpheus hemphilli Coutière, 1909. Photo by Arthur Anker. Synalpheus iphinoe De Man, 1909a = Synalpheus Iphinoë De Man, 1909a: 116. [8°23'.5S 119°4'.6E, Sapeh-strait, 70 m; Madura-bay and other localities in the southern part of Molo-strait, 54-90 m; Banda-anchorage, 9-36 m; Rumah-ku- da-bay, Roma-island, 36 m] Synalpheus iocasta De Man, 1909a = Synalpheus Iocasta De Man, 1909a: 119. [Makassar and surroundings, up to 32 m; 0°58'.5N 122°42'.5E, west of Kwadang-bay-entrance, 72 m; Anchorage north of Salomakiëe (Damar) is- land, 45 m; 1°42'.5S 130°47'.5E, 32 m; 4°20'S 122°58'E, between islands of Wowoni and Buton, northern entrance of Buton-strait, 75-94 m; Banda-anchorage, 9-36 m; Anchorage off Pulu Jedan, east coast of Aru-islands (Pearl-banks), 13 m; 5°28'.2S 134°53'.9E, 57 m; 8°25'.2S 127°18'.4E, an- chorage between Nusa Besi and the N.E. point of Timor, 27-54 m; 8°39'.1 127°4'.4E, anchorage south coast of Timor, 34 m; Mid-channel in Solor-strait off Kampong Menanga, 113 m; 8°30'S 119°7'.5E, 73 m] Synalpheus irie MacDonald, Hultgren & Duffy, 2009: 25; Figs 11-16; Plate 3C-D. [fore-reef (near M1 chan- nel marker), 18°28.083'N 77°23.289'W, from canals of Auletta cf. sycinularia] Synalpheus jedanensis De Man, 1909a: 117. [Anchorage off Pulu Jedan, east coast of Aru-islands (Pearl- banks), 13 m] Synalpheus kensleyi (Ríos & Duffy, 2007) = Zuzalpheus kensleyi Ríos & Duffy, 2007: 41; Figs 18-22; Plate 3. -

South China Tiger Conservation Strategy
South China Tiger Conservation Strategy Petri Viljoen Objective The South China Tiger Conservation Strategy aims to establish genetically viable, free-ranging South China tigers in suitable, newly created protected areas in south- eastern China. Background The South China Tiger ( Panthera tigris amoyensis ), also known as the Chinese Tiger, is the most endangered of the remaining living tiger subspecies (IUCN 2009). It is estimated that a small number (10-20) of free-ranging South China tigers could still be present, although no recent sightings of this tiger subspecies in its natural range have been reported. The captive population, all in Chinese zoos with the exception of 14 individuals currently in South Africa, number approximately 60 (Traylor-Holzer 1998; IUCN 2009). The South China Tiger is considered to be critically endangered (IUCN 2009) and precariously close to extinction (Tilson et al. 1997; Tilson et al. 2004). Herrington (1987) considered the South China Tiger to be a relic population of the "stem" tiger, possibly occurring close to the area of the species’ origin. Luo et al. (2004, in IUCN 2009) suggested that the centre of the original tiger radiation was most likely the southern China/northern Indochina region. Save China’s Tigers (SCT) is a UK, US, China and Hong Kong based charitable foundation. SCT’s specific mission is to save the South China Tiger from extinction and recreate a viable population of free-ranging South China Tigers in protected reserves in their former range. To achieve this objective SCT aims to assist with the development of captive breeding and reintroduction of South China Tigers in China. -

Tulane Law School
Herbert Larson Executive Director for International Legal Programs Maria Landry Director of Admission and International Student Recruitment and Enrollment Mallory Asp Senior Administrative Coordinator International Legal Programs 2018 - 2019 International Exchange Programs: Semester Abroad WHO: Any qualified J.D. student may apply to go abroad in the second semester of 2L year, or in the first semester of 3L year. WHAT: An academic exchange program with law schools outside the United States. Tulane students live and study abroad for one semester and earn credit (up to 14 credits on a pass/fail basis) towards the Tulane J.D. The programs relate to the socio-legal environment of the hosting country and/or have an international or comparative law focus. WHERE: Tulane has exchange programs in numerous countries and languages: Programs in English: − University of Amsterdam − Bucerius University (Hamburg) − University of Copenhagen − University of Hong Kong − University of New South Wales (Sydney) − Utrecht University − Tel Aviv University − Stockholm University Programs with Mixed Language Offerings: Programs in Language of Country: − China University of Political Science and Law − Università di Bologna (Beijing) − University of Buenos Aires − ESADE Law School (Barcelona) − Université de Strasbourg − Fudan University (Shanghai) − Universidad Autónoma de Nuevo León − Universidad de Carlos III (Madrid) − Università di Siena − Tsinghua University (Beijing) − Dalian Maritime University − Universidad de los Andes (Bogotá) − University of Zurich (Most courses in German) WHY: Well-qualified students interested in international or comparative law can gain firsthand experience with foreign procedures, laws, practice, education, and culture. WHEN: Exchange programs run in the fall and spring semesters. Each have separate application deadlines. -

HIDDEN in PLAIN SIGHT China's Clandestine Tiger Trade ACKNOWLEDGEMENTS CONTENTS
HIDDEN IN PLAIN SIGHT China's Clandestine Tiger Trade ACKNOWLEDGEMENTS CONTENTS This report was written by Environmental Investigation Agency. 1 EIA would like to thank the Rufford Foundation, SUMMARY the David Shepherd Wildlife Foundation, Ernest Kleinwort Charitable Trust and Save Wild Tigers for their support in making this work possible. 3 INTRODUCTION Special thanks to our colleagues “on the frontline” in tiger range countries for their information, 5 advice and inspiration. LEGAL CONTEXT ® EIA uses IBM i2 intelligence analysis software. 6 INVESTIGATION FINDINGS Report design by: www.designsolutions.me.uk 12 TIGER FARMING February 2013 13 PREVIOUS EXPOSÉS 17 STIMULATING POACHING OF WILD ASIAN BIG CATS 20 MIXED MESSAGES 24 SOUTH-EAST ASIA TIGER FARMS 25 CONCLUSIONS AND RECOMMENDATIONS 26 APPENDICES 1) TABLE OF LAWS AND REGULATIONS RELATING TO TIGERS IN CHINA 2) TIGER FARMING TIMELINE ENVIRONMENTAL INVESTIGATION AGENCY (EIA) 62/63 Upper Street, London N1 0NY, UK Tel: +44 (0) 20 7354 7960 Fax: +44 (0) 20 7354 7961 email: [email protected] www.eia-international.org EIA US P.O.Box 53343 Washington DC 20009 USA Tel: +1 202 483 6621 Fax: +1 202 986 8626 email: [email protected] www.eia-global.org Front cover image © Robin Hamilton All images © EIA unless otherwise specified © Michael Vickers/www.tigersintheforest.co.uk SUMMARY Undercover investigations and a review of available Chinese laws have revealed that while China banned tiger bone trade for medicinal uses in 1993, it has encouraged the growth of the captive-breeding of tigers to supply a quietly expanding legal domestic trade in tiger skins.