Deep Learning for Computational Science and Engineering Abstract
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
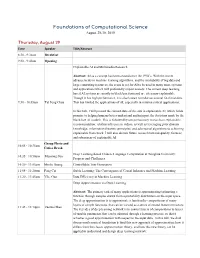
Foundations of Computational Science August 29-30, 2019
Foundations of Computational Science August 29-30, 2019 Thursday, August 29 Time Speaker Title/Abstract 8:30 - 9:30am Breakfast 9:30 - 9:40am Opening Explainable AI and Multimedia Research Abstract: AI as a concept has been around since the 1950’s. With the recent advancements in machine learning algorithms, and the availability of big data and large computing resources, the scene is set for AI to be used in many more systems and applications which will profoundly impact society. The current deep learning based AI systems are mostly in black box form and are often non-explainable. Though it has high performance, it is also known to make occasional fatal mistakes. 9:30 - 10:05am Tat Seng Chua This has limited the applications of AI, especially in mission critical applications. In this talk, I will present the current state-of-the arts in explainable AI, which holds promise to helping humans better understand and interpret the decisions made by the black-box AI models. This is followed by our preliminary research on explainable recommendation, relation inference in videos, as well as leveraging prior domain knowledge, information theoretic principles, and adversarial algorithms to achieving explainable framework. I will also discuss future research towards quality, fairness and robustness of explainable AI. Group Photo and 10:05 - 10:35am Coffee Break Deep Learning-based Chinese Language Computation at Tsinghua University: 10:35 - 10:50am Maosong Sun Progress and Challenges 10:50 - 11:05am Minlie Huang Controllable Text Generation 11:05 - 11:20am Peng Cui Stable Learning: The Convergence of Causal Inference and Machine Learning 11:20 - 11:45am Yike Guo Data Efficiency in Machine Learning Deep Approximation via Deep Learning Abstract: The primary task of many applications is approximating/estimating a function through samples drawn from a probability distribution on the input space. -

Computational Science and Engineering
Computational Science and Engineering, PhD College of Engineering Graduate Coordinator: TBA Email: Phone: Department Chair: Marwan Bikdash Email: [email protected] Phone: 336-334-7437 The PhD in Computational Science and Engineering (CSE) is an interdisciplinary graduate program designed for students who seek to use advanced computational methods to solve large problems in diverse fields ranging from the basic sciences (physics, chemistry, mathematics, etc.) to sociology, biology, engineering, and economics. The mission of Computational Science and Engineering is to graduate professionals who (a) have expertise in developing novel computational methodologies and products, and/or (b) have extended their expertise in specific disciplines (in science, technology, engineering, and socioeconomics) with computational tools. The Ph.D. program is designed for students with graduate and undergraduate degrees in a variety of fields including engineering, chemistry, physics, mathematics, computer science, and economics who will be trained to develop problem-solving methodologies and computational tools for solving challenging problems. Research in Computational Science and Engineering includes: computational system theory, big data and computational statistics, high-performance computing and scientific visualization, multi-scale and multi-physics modeling, computational solid, fluid and nonlinear dynamics, computational geometry, fast and scalable algorithms, computational civil engineering, bioinformatics and computational biology, and computational physics. Additional Admission Requirements Master of Science or Engineering degree in Computational Science and Engineering (CSE) or in science, engineering, business, economics, technology or in a field allied to computational science or computational engineering field. GRE scores Program Outcomes: Students will demonstrate critical thinking and ability in conducting research in engineering, science and mathematics through computational modeling and simulations. -

Artificial Intelligence Research For
Charles Xie ([email protected]) is a senior scientist who works at the inter- section between computational science and learning science. Artificial Intelligence research for engineering design By Charles Xie Imagine a high school classroom in 2030. AI is one of the three most promising areas for which Bill Gates Students are challenged to design an said earlier this year he would drop out of college again if he were a young student today. The victory of Google’s AlphaGo against engineering system that meets the needs of a top human Go player in 2016 was a landmark in the history of their community. They work with advanced AI. The game of Go is considered a difficult challenge because computer-aided design (CAD) software that the number of legal positions on a 19×19 grid is estimated to be leverages the power of scientific simulation, 2×10170, far more than the total number of atoms in the observ- mixed reality, and cloud computing. Every able universe combined (1082). While AlphaGo may be only a cool tool needed to complete an engineering type of weak AI that plays Go at present, scientists believe that its development will engender technologies that eventually find design project is seamlessly integrated into applications in other fields. the CAD platform to streamline the learning process. But the most striking capability of Performance modeling the software is its ability to act like a mentor (e.g., simulation-based analysis) who has all the knowledge about the design project to answer questions, learns from all the successful or unsuccessful solutions ever Evaluator attempted by human designers or computer agents, adapts to the needs of each student based on experience interacting with all sorts of designers, inspires students to explore creative ideas, and, most importantly, remains User Ideator responsive all the time without ever running out of steam. -

An Evaluation of Tensorflow As a Programming Framework for HPC Applications
DEGREE PROJECT IN COMPUTER SCIENCE AND ENGINEERING, SECOND CYCLE, 30 CREDITS STOCKHOLM, SWEDEN 2018 An Evaluation of TensorFlow as a Programming Framework for HPC Applications WEI DER CHIEN KTH ROYAL INSTITUTE OF TECHNOLOGY SCHOOL OF ELECTRICAL ENGINEERING AND COMPUTER SCIENCE An Evaluation of TensorFlow as a Programming Framework for HPC Applications WEI DER CHIEN Master in Computer Science Date: August 28, 2018 Supervisor: Stefano Markidis Examiner: Erwin Laure Swedish title: En undersökning av TensorFlow som ett utvecklingsramverk för högpresterande datorsystem School of Electrical Engineering and Computer Science iii Abstract In recent years, deep-learning, a branch of machine learning gained increasing popularity due to their extensive applications and perfor- mance. At the core of these application is dense matrix-matrix multipli- cation. Graphics Processing Units (GPUs) are commonly used in the training process due to their massively parallel computation capabili- ties. In addition, specialized low-precision accelerators have emerged to specifically address Tensor operations. Software frameworks, such as TensorFlow have also emerged to increase the expressiveness of neural network model development. In TensorFlow computation problems are expressed as Computation Graphs where nodes of a graph denote operation and edges denote data movement between operations. With increasing number of heterogeneous accelerators which might co-exist on the same cluster system, it became increasingly difficult for users to program efficient and scalable applications. TensorFlow provides a high level of abstraction and it is possible to place operations of a computation graph on a device easily through a high level API. In this work, the usability of TensorFlow as a programming framework for HPC application is reviewed. -

A Brief Introduction to Mathematical Optimization in Julia (Part 1) What Is Julia? and Why Should I Care?
A Brief Introduction to Mathematical Optimization in Julia (Part 1) What is Julia? And why should I care? Carleton Coffrin Advanced Network Science Initiative https://lanl-ansi.github.io/ Managed by Triad National Security, LLC for the U.S. Department of Energy’s NNSA LA-UR-21-20278 Warning: This is not a technical talk! 9/25/2019 Julia_prog_language.svg What is Julia? • A “new” programming language developed at MIT • nearly 10 years old (appeared around 2012) • Julia is a high-level, high-performance, dynamic programming language • free and open source • The Creator’s Motivation • build the next-generation of programming language for numerical analysis and computational science • Solve the “two-language problem” (you havefile:///Users/carleton/Downloads/Julia_prog_language.svg to choose between 1/1 performance and implementation simplicity) Who is using Julia? Julia is much more than an academic curiosity! See https://juliacomputing.com/ Why Julia? Application9/25/2019 Julia_prog_language.svg Needs Data Scince Numerical Scripting Computing Automation 9/25/2019 Thin-Border-Logo-Text Julia Data Differential Equations file:///Users/carleton/Downloads/Julia_prog_language.svg 1/1 Deep Learning Mathematical Optimization High Performance https://camo.githubusercontent.com/31d60f762b44d0c3ea47cc16b785e042104a6e03/68747470733a2f2f7777772e6a756c69616f70742e6f72672f696d616765732f6a7… 1/1 -

From Computational Science to Science Discovery
FromComputationalSciencetoScienceDiscovery:The NextComputingLandscape GiladShainer,BrianSparks,ScotSchultz,EricLantz,WilliamLiu,TongLiu,GoldiMisra HPCAdvisoryCouncil {Gilad,Brian,Scot,Eric,William,Tong,[email protected]} Computationalscienceisthefieldofstudyconcernedwithconstructingmathematicalmodelsand numericaltechniquesthatrepresentscientific,socialscientificorengineeringproblemsandemploying thesemodelsoncomputers,orclustersofcomputerstoanalyze,exploreorsolvethesemodels. Numericalsimulationenablesthestudyofcomplexphenomenathatwouldbetooexpensiveor dangeroustostudybydirectexperimentation.Thequestforeverhigherlevelsofdetailandrealismin suchsimulationsrequiresenormouscomputationalcapacity,andhasprovidedtheimpetusfor breakthroughsincomputeralgorithmsandarchitectures.Duetotheseadvances,computational scientistsandengineerscannowsolvelargeͲscaleproblemsthatwereoncethoughtintractableby creatingtherelatedmodelsandsimulatethemviahighͲperformancecomputeclustersor supercomputers.Simulationisbeingusedasanintegralpartofthemanufacturing,designanddecisionͲ makingprocesses,andasafundamentaltoolforscientificresearch.ProblemswherehighͲperformance simulationplayapivotalroleincludeforexampleweatherandclimateprediction,nuclearandenergy research,simulationanddesignofvehiclesandaircrafts,electronicdesignautomation,astrophysics, quantummechanics,biology,computationalchemistryandmore. Computationalscienceiscommonlyconsideredthethirdmodeofscience,wherethepreviousmodesor paradigmswereexperimentation/observationandtheory.Inthepast,sciencewasperformedby -

Julia: a Fresh Approach to Numerical Computing∗
SIAM REVIEW c 2017 Society for Industrial and Applied Mathematics Vol. 59, No. 1, pp. 65–98 Julia: A Fresh Approach to Numerical Computing∗ Jeff Bezansony Alan Edelmanz Stefan Karpinskix Viral B. Shahy Abstract. Bridging cultures that have often been distant, Julia combines expertise from the diverse fields of computer science and computational science to create a new approach to numerical computing. Julia is designed to be easy and fast and questions notions generally held to be \laws of nature" by practitioners of numerical computing: 1. High-level dynamic programs have to be slow. 2. One must prototype in one language and then rewrite in another language for speed or deployment. 3. There are parts of a system appropriate for the programmer, and other parts that are best left untouched as they have been built by the experts. We introduce the Julia programming language and its design|a dance between special- ization and abstraction. Specialization allows for custom treatment. Multiple dispatch, a technique from computer science, picks the right algorithm for the right circumstance. Abstraction, which is what good computation is really about, recognizes what remains the same after differences are stripped away. Abstractions in mathematics are captured as code through another technique from computer science, generic programming. Julia shows that one can achieve machine performance without sacrificing human con- venience. Key words. Julia, numerical, scientific computing, parallel AMS subject classifications. 68N15, 65Y05, 97P40 DOI. 10.1137/141000671 Contents 1 Scientific Computing Languages: The Julia Innovation 66 1.1 Julia Architecture and Language Design Philosophy . 67 ∗Received by the editors December 18, 2014; accepted for publication (in revised form) December 16, 2015; published electronically February 7, 2017. -

COMPUTATIONAL SCIENCE 2017-2018 College of Engineering and Computer Science BACHELOR of SCIENCE Computer Science
COMPUTATIONAL SCIENCE 2017-2018 College of Engineering and Computer Science BACHELOR OF SCIENCE Computer Science *The Computational Science program offers students the opportunity to acquire a knowledge in computing integrated with knowledge in one of the following areas of study: (a) bioinformatics, (b) computational physics, (c) computational chemistry, (d) computational mathematics, (e) environmental science informatics, (f) health informatics, (g) digital forensics and cyber security, (h) business informatics, (i) biomedical informatics, (j) computational engineering physics, and (k) computational engineering technology. Graduates of this program major in computational science with a concentration in one of the above areas of study. (Amended for clarification 12/5/2018). *Previous UTRGV language: Computational science graduates develop emphasis in two major fields, one in computer science and one in another field, in order to integrate an interdisciplinary computing degree applied to a number of emerging areas of study such as biomedical-informatics, digital forensics, computational chemistry, and computational physics, to mention a few examples. Graduates of this program are prepared to enter the workforce or to continue a graduate studies either in computer science or in the second major. A – GENERAL EDUCATION CORE – 42 HOURS Students must fulfill the General Education Core requirements. The courses listed below satisfy both degree requirements and General Education core requirements. Required 020 - Mathematics – 3 hours For all -

Machine Learning for Synchronized Swimming
Machine Learning for Synchronised Swimming Siddhartha Verma, Guido Novati Petros Koumoutsakos FSIM 2017 May 25, 2017 CSElab Computational Science & Engineering Laboratory http://www.cse-lab.ethz.ch Motivation • Why do fish swim in groups? • Social behaviour? • Anti-predator benefits • Better foraging/reproductive opportunities • Better problem-solving capability • Propulsive advantage? Credit: Artbeats • Classical works on schooling: Breder (1965), Weihs (1973,1975), … • Based on simplified inviscid models (e.g., point vortices) • More recent simulations: consider pre-assigned and fixed formations Hemelrijk et al. (2015), Daghooghi & Borazjani (2015) • But: actual formations evolve dynamically Breder (1965) Weihs (1973), Shaw (1978) • Essential question: Possible to swim autonomously & energetically favourable? Hemelrijk et al. (2015) Vortices in the wake • Depending on Re and swimming- kinematics, double row of vortex rings in the wake • Our goal: exploit the flow generated by these vortices Numerical methods 0 in 2D • Remeshed vortex methods (2D) @! 2 + u ! = ! u + ⌫ ! + λ (χ (us u)) • Solve vorticity form of incompressible Navier-Stokes @t ·r ·r r r⇥ − Advection Diffusion Penalization • Brinkman penalization • Accounts for fluid-solid interaction Angot et al., Numerische Mathematik (1999) • 2D: Wavelet-based adaptive grid • Cost-effective compared to uniform grids Rossinelli et al., J. Comput. Phys. (2015) • 3D: Uniform grid Finite Volume solver Rossinelli et al., SC'13 Proc. Int. Conf. High Perf. Comput., Denver, Colorado Interacting swimmers -

Section Computerscience.Pdf
Computer Science | 27 Ganesh Baliga Professor Computer Science [email protected] http://elvis.rowan.edu/~baliga/research.pdf Education: B. Tech. (Computer Science and Engineering), Indian Institute of Technology, Bombay M. Tech. (Computer Science and Engineering), Indian Institute of Technology, Bombay MS (Computer and Information Sciences), University of Delaware PhD (Computer and Information Sciences), University of Delaware Research Expertise: Data Analytics | Machine Learning | Algorithm Design and Analysis | Cloud Computing My research focus is on the design of algorithms and software systems for machine learning and data analytics. I have published over 15 papers in machine learning in international conferences and journals. Presently, I am the Technical Lead and co-PI of the Perka First Data-Rowan CS Lab, an innovative industry academia collaboration where Perka engineers work closely with faculty and students to develop active production software. At present I am establishing a lab with a focus on data analytics using deep neural networks. I am the co-PI of a grant from the National Science Foundation to develop materials for an undergraduate curriculum for algorithms design and NP-Completeness. Over the past four years, I have served as co-PI in projects funded by Bristol-Myers Squibb and Mission Solutions Engineering and have been involved in nine external grants and contracts. Member of: ACM Recent Academic Projects: Co-PI, Perka Lab. Sponsored by Perka Inc., May 2015 – May 2018. Co-PI, NSF TUES grant: “Learning algorithm design: A project based curriculum” May 2012 – April 2017. Recent Publications: Lobo AF, Baliga GR (2017) A project-based curriculum for algorithm design and NP completeness centered on the Sudoku problem. -

Verification and Validation in Computational Fluid Dynamics1
SAND2002 - 0529 Unlimited Release Printed March 2002 Verification and Validation in Computational Fluid Dynamics1 William L. Oberkampf Validation and Uncertainty Estimation Department Timothy G. Trucano Optimization and Uncertainty Estimation Department Sandia National Laboratories P. O. Box 5800 Albuquerque, New Mexico 87185 Abstract Verification and validation (V&V) are the primary means to assess accuracy and reliability in computational simulations. This paper presents an extensive review of the literature in V&V in computational fluid dynamics (CFD), discusses methods and procedures for assessing V&V, and develops a number of extensions to existing ideas. The review of the development of V&V terminology and methodology points out the contributions from members of the operations research, statistics, and CFD communities. Fundamental issues in V&V are addressed, such as code verification versus solution verification, model validation versus solution validation, the distinction between error and uncertainty, conceptual sources of error and uncertainty, and the relationship between validation and prediction. The fundamental strategy of verification is the identification and quantification of errors in the computational model and its solution. In verification activities, the accuracy of a computational solution is primarily measured relative to two types of highly accurate solutions: analytical solutions and highly accurate numerical solutions. Methods for determining the accuracy of numerical solutions are presented and the importance of software testing during verification activities is emphasized. The fundamental strategy of 1Accepted for publication in the review journal Progress in Aerospace Sciences. 3 validation is to assess how accurately the computational results compare with the experimental data, with quantified error and uncertainty estimates for both. -

Computational Physics * Cruz a Editorial Office, Pure and Applied Physics, India
Research & Reviews: Journal of Pure and Applied Physics e-ISSN:2320-2459 p-ISSN:2347-2316 Computational Physics * Cruz A Editorial Office, Pure and Applied Physics, India Editorial Note Received date: 21/02/2021 *For Correspondence Accepted date: 23/02/2021 Published date: 28/02/2021 Andrew Cruz, Computational Physics, Editorial Office, Pure and Applied Physics, India. E-mail: [email protected] EDITORIAL NOTE Computational physics is that the study and implementation of numerical analysis to unravel problems in physics that a quantitative theory already exists. Historically, computational physics was the primary application of recent computers in science, and is now a subset of computational science. It is sometimes considered a sub discipline (or offshoot) of theoretical physics, but others consider it an intermediate branch between theoretical and experimental physics-a neighbourhood of study which supplements both theory and experiment. Computational physics problems are generally very difficult to unravel exactly. This is often thanks to several (mathematical) reasons: lack of algebraic and/or analytic solvability, complexity, and chaos. For example, even apparently simple problems, like calculating the wave function of an electron orbiting an atom during a strong field (Stark effect), may require great effort to formulate a practical algorithm (if one are often found); other cruder or brute-force techniques, like graphical methods or root finding, could also be required. On the more advanced side, mathematical perturbation theory is additionally sometimes used (a working is shown for this particular example here). In addition, the computational cost and computational complexity for many-body problems (and their classical counterparts) tend to grow quickly.