A Genomics-Based Approach to Biodefence Preparedness
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Characterization of the Interaction Between R. Conorii and Human
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 4-5-2018 Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis Abigail Inez Fish Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Bacteria Commons, Bacteriology Commons, Biology Commons, Immunology of Infectious Disease Commons, and the Pathogenic Microbiology Commons Recommended Citation Fish, Abigail Inez, "Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis" (2018). LSU Doctoral Dissertations. 4566. https://digitalcommons.lsu.edu/gradschool_dissertations/4566 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. CHARACTERIZATION OF THE INTERACTION BETWEEN R. CONORII AND HUMAN HOST VITRONECTIN IN RICKETTSIAL PATHOGENESIS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Interdepartmental Program in Biomedical and Veterinary Medical Sciences Through the Department of Pathobiological Sciences by Abigail Inez -

BD-CS-057, REV 0 | AUGUST 2017 | Page 1
EXPLIFY RESPIRATORY PATHOGENS BY NEXT GENERATION SEQUENCING Limitations Negative results do not rule out viral, bacterial, or fungal infections. Targeted, PCR-based tests are generally more sensitive and are preferred when specific pathogens are suspected, especially for DNA viruses (Adenovirus, CMV, HHV6, HSV, and VZV), mycobacteria, and fungi. The analytical sensitivity of this test depends on the cellularity of the sample and the concentration of all microbes present. Analytical sensitivity is assessed using Internal Controls that are added to each sample. Sequencing data for Internal Controls is quantified. Samples with Internal Control values below the validated minimum may have reduced analytical sensitivity or contain inhibitors and are reported as ‘Reduced Analytical Sensitivity’. Additional respiratory pathogens to those reported cannot be excluded in samples with ‘Reduced Analytical Sensitivity’. Due to the complexity of next generation sequencing methodologies, there may be a risk of false-positive results. Contamination with organisms from the upper respiratory tract during specimen collection can also occur. The detection of viral, bacterial, and fungal nucleic acid does not imply organisms causing invasive infection. Results from this test need to be interpreted in conjunction with the clinical history, results of other laboratory tests, epidemiologic information, and other available data. Confirmation of positive results by an alternate method may be indicated in select cases. Validated Organisms BACTERIA Achromobacter -

Phenotypic and Genomic Analyses of Burkholderia Stabilis Clinical Contamination, Switzerland Helena M.B
RESEARCH Phenotypic and Genomic Analyses of Burkholderia stabilis Clinical Contamination, Switzerland Helena M.B. Seth-Smith, Carlo Casanova, Rami Sommerstein, Dominik M. Meinel,1 Mohamed M.H. Abdelbary,2 Dominique S. Blanc, Sara Droz, Urs Führer, Reto Lienhard, Claudia Lang, Olivier Dubuis, Matthias Schlegel, Andreas Widmer, Peter M. Keller,3 Jonas Marschall, Adrian Egli A recent hospital outbreak related to premoistened gloves pathogens that generally fall within the B. cepacia com- used to wash patients exposed the difficulties of defining plex (Bcc) (1). Burkholderia bacteria have large, flexible, Burkholderia species in clinical settings. The outbreak strain multi-replicon genomes, a large metabolic repertoire, vari- displayed key B. stabilis phenotypes, including the inabil- ous virulence factors, and inherent resistance to many anti- ity to grow at 42°C; we used whole-genome sequencing to microbial drugs (2,3). confirm the pathogen was B. stabilis. The outbreak strain An outbreak of B. stabilis was identified among hos- genome comprises 3 chromosomes and a plasmid, shar- ing an average nucleotide identity of 98.4% with B. stabilis pitalized patients across several cantons in Switzerland ATCC27515 BAA-67, but with 13% novel coding sequenc- during 2015–2016 (4). The bacterium caused bloodstream es. The genome lacks identifiable virulence factors and has infections, noninvasive infections, and wound contamina- no apparent increase in encoded antimicrobial drug resis- tions. The source of the infection was traced to contaminat- tance, few insertion sequences, and few pseudogenes, ed commercially available, premoistened washing gloves suggesting this outbreak was an opportunistic infection by used for bedridden patients. After hospitals discontinued an environmental strain not adapted to human pathogenic- use of these gloves, the outbreak resolved. -

“Candidatus Deianiraea Vastatrix” with the Ciliate Paramecium Suggests
bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The extracellular association of the bacterium “Candidatus Deianiraea vastatrix” with the ciliate Paramecium suggests an alternative scenario for the evolution of Rickettsiales 5 Castelli M.1, Sabaneyeva E.2, Lanzoni O.3, Lebedeva N.4, Floriano A.M.5, Gaiarsa S.5,6, Benken K.7, Modeo L. 3, Bandi C.1, Potekhin A.8, Sassera D.5*, Petroni G.3* 1. Centro Romeo ed Enrica Invernizzi Ricerca Pediatrica, Dipartimento di Bioscienze, Università 10 degli studi di Milano, Milan, Italy 2. Department of Cytology and Histology, Faculty of Biology, Saint Petersburg State University, Saint-Petersburg, Russia 3. Dipartimento di Biologia, Università di Pisa, Pisa, Italy 4 Centre of Core Facilities “Culture Collections of Microorganisms”, Saint Petersburg State 15 University, Saint Petersburg, Russia 5. Dipartimento di Biologia e Biotecnologie, Università degli studi di Pavia, Pavia, Italy 6. UOC Microbiologia e Virologia, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 7. Core Facility Center for Microscopy and Microanalysis, Saint Petersburg State University, Saint- Petersburg, Russia 20 8. Department of Microbiology, Faculty of Biology, Saint Petersburg State University, Saint- Petersburg, Russia * Corresponding authors, contacts: [email protected] ; [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -

Mechanisms of Rickettsia Parkeri Invasion of Host Cells and Early Actin-Based Motility
Mechanisms of Rickettsia parkeri invasion of host cells and early actin-based motility By Shawna Reed A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Microbiology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Matthew D. Welch, Chair Professor David G. Drubin Professor Daniel Portnoy Professor Kathleen R. Ryan Fall 2012 Mechanisms of Rickettsia parkeri invasion of host cells and early actin-based motility © 2012 By Shawna Reed ABSTRACT Mechanisms of Rickettsia parkeri invasion of host cells and early actin-based motility by Shawna Reed Doctor of Philosophy in Microbiology University of California, Berkeley Professor Matthew D. Welch, Chair Rickettsiae are obligate intracellular pathogens that are transmitted to humans by arthropod vectors and cause diseases such as spotted fever and typhus. Spotted fever group (SFG) Rickettsia hijack the host actin cytoskeleton to invade, move within, and spread between eukaryotic host cells during their obligate intracellular life cycle. Rickettsia express two bacterial proteins that can activate actin polymerization: RickA activates the host actin-nucleating Arp2/3 complex while Sca2 directly nucleates actin filaments. In this thesis, I aimed to resolve which host proteins were required for invasion and intracellular motility, and to determine how the bacterial proteins RickA and Sca2 contribute to these processes. Although rickettsiae require the host cell actin cytoskeleton for invasion, the cytoskeletal proteins that mediate this process have not been completely described. To identify the host factors important during cell invasion by Rickettsia parkeri, a member of the SFG, I performed an RNAi screen targeting 105 proteins in Drosophila melanogaster S2R+ cells. -

Gene Gain and Loss Events in Rickettsia and Orientia Species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2*
Georgiades et al. Biology Direct 2011, 6:6 http://www.biology-direct.com/content/6/1/6 RESEARCH Open Access Gene gain and loss events in Rickettsia and Orientia species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2* Abstract Background: Genome degradation is an ongoing process in all members of the Rickettsiales order, which makes these bacterial species an excellent model for studying reductive evolution through interspecies variation in genome size and gene content. In this study, we evaluated the degree to which gene loss shaped the content of some Rickettsiales genomes. We shed light on the role played by horizontal gene transfers in the genome evolution of Rickettsiales. Results: Our phylogenomic tree, based on whole-genome content, presented a topology distinct from that of the whole core gene concatenated phylogenetic tree, suggesting that the gene repertoires involved have different evolutionary histories. Indeed, we present evidence for 3 possible horizontal gene transfer events from various organisms to Orientia and 6 to Rickettsia spp., while we also identified 3 possible horizontal gene transfer events from Rickettsia and Orientia to other bacteria. We found 17 putative genes in Rickettsia spp. that are probably the result of de novo gene creation; 2 of these genes appear to be functional. On the basis of these results, we were able to reconstruct the gene repertoires of “proto-Rickettsiales” and “proto-Rickettsiaceae”, which correspond to the ancestors of Rickettsiales and Rickettsiaceae, respectively. Finally, we found that 2,135 genes were lost during the evolution of the Rickettsiaceae to an intracellular lifestyle. Conclusions: Our phylogenetic analysis allowed us to track the gene gain and loss events occurring in bacterial genomes during their evolution from a free-living to an intracellular lifestyle. -

Rickettsial Infections and Their Clinical Presentations in the Western Province of Sri Lanka: a Hospital-Based Study§
International Journal of Infectious Diseases (2008) 12, 198—202 http://intl.elsevierhealth.com/journals/ijid Rickettsial infections and their clinical presentations in the Western Province of Sri Lanka: a hospital-based study§ R. Premaratna a,*, A.D. Loftis b, T.G.A.N. Chandrasena c, G.A. Dasch b, H.J. de Silva a a Department of Medicine, Faculty of Medicine, University of Kelaniya, PO Box 6, Thalagolla Rd, Ragama, Sri Lanka b Viral and Rickettsial Zoonoses Branch, Centers for Disease Control and Prevention, Atlanta, GA 30333, USA c Department of Parasitology, Faculty of Medicine, University of Kelaniya, Sri Lanka Received 18 September 2006; received in revised form 21 February 2007; accepted 18 June 2007 Corresponding Editor: Craig Lee, Ottawa, Canada KEYWORDS Summary Scrub typhus; Background: Rickettsial infections are re-emerging. A study of the geographical distribution of Spotted fever group rickettsial infections, their clinical manifestations, and their complications would facilitate early rickettsioses; diagnosis. Sri Lanka Methods: Thirty-one selected patients from the Western Province of Sri Lanka were studied for rickettsial species, clinical manifestations, and complications. Results: Of 31 patients with possible rickettsioses, 29 (94%) fell into the categories of confirmed, presumptive, or exposed cases of acute rickettsial infections (scrub typhus was diagnosed in 19 (66%), spotted fever group in eight (28%)). Early acute infection or past exposure was suggested in two (7%) cases; cross-reactivity of antigens or past exposure to one or more species was suggested in nine (31%). Seventeen out of 19 (89%) patients with scrub typhus had eschars. Nine out of 29 (32%) patients had a discrete erythematous papular rash: seven caused by spotted fever group, two by scrub typhus. -

Insights Into the Pathogenicity of Burkholderia Pseudomallei
REVIEWS Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei W. Joost Wiersinga*, Tom van der Poll*, Nicholas J. White‡§, Nicholas P. Day‡§ and Sharon J. Peacock‡§ Abstract | Burkholderia pseudomallei is a potential bioterror agent and the causative agent of melioidosis, a severe disease that is endemic in areas of Southeast Asia and Northern Australia. Infection is often associated with bacterial dissemination to distant sites, and there are many possible disease manifestations, with melioidosis septic shock being the most severe. Eradication of the organism following infection is difficult, with a slow fever-clearance time, the need for prolonged antibiotic therapy and a high rate of relapse if therapy is not completed. Mortality from melioidosis septic shock remains high despite appropriate antimicrobial therapy. Prevention of disease and a reduction in mortality and the rate of relapse are priority areas for future research efforts. Studying how the disease is acquired and the host–pathogen interactions involved will underpin these efforts; this review presents an overview of current knowledge in these areas, highlighting key topics for evaluation. Melioidosis is a serious disease caused by the aerobic, rifamycins, colistin and aminoglycosides), but is usually Gram-negative soil-dwelling bacillus Burkholderia pseu- susceptible to amoxicillin-clavulanate, chloramphenicol, domallei and is most common in Southeast Asia and doxycycline, trimethoprim-sulphamethoxazole, ureido- Northern Australia. Melioidosis is responsible for 20% of penicillins, ceftazidime and carbapenems2,4. Treatment all community-acquired septicaemias and 40% of sepsis- is required for 20 weeks and is divided into intravenous related mortality in northeast Thailand. Reported cases are and oral phases2,4. Initial intravenous therapy is given likely to represent ‘the tip of the iceberg’1,2, as confirmation for 10–14 days; ceftazidime or a carbapenem are the of disease depends on bacterial isolation, a technique that drugs of choice. -
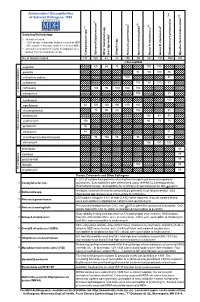
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire