Watch the Presentation Here
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Late Holocene Sea Level Rise in Southwest Florida: Implications for Estuarine Management and Coastal Evolution
LATE HOLOCENE SEA LEVEL RISE IN SOUTHWEST FLORIDA: IMPLICATIONS FOR ESTUARINE MANAGEMENT AND COASTAL EVOLUTION Dana Derickson, Figure 2 FACULTY Lily Lowery, University of the South Mike Savarese, Florida Gulf Coast University Stephanie Obley, Flroida Gulf Coast University Leonre Tedesco, Indiana University and Purdue Monica Roth, SUNYOneonta University at Indianapolis Ramon Lopez, Vassar College Carol Mankiewcz, Beloit College Lora Shrake, TA, Indiana University and Purdue University at Indianapolis VISITING and PARTNER SCIENTISTS Gary Lytton, Michael Shirley, Judy Haner, STUDENTS Leslie Breland, Dave Liccardi, Chuck Margo Burton, Whitman College McKenna, Steve Theberge, Pat O’Donnell, Heather Stoffel, Melissa Hennig, and Renee Dana Derickson, Trinity University Wilson, Rookery Bay NERR Leda Jackson, Indiana University and Purdue Joe Kakareka, Aswani Volety, and Win University at Indianapolis Everham, Florida Gulf Coast University Chris Kitchen, Whitman College Beth A. Palmer, Consortium Coordinator Nicholas Levsen, Beloit College Emily Lindland, Florida Gulf Coast University LATE HOLOCENE SEA LEVEL RISE IN SOUTHWEST FLORIDA: IMPLICATIONS FOR ESTUARINE MANAGEMENT AND COASTAL EVOLUTION MICHAEL SAVARESE, Florida Gulf Coast University LENORE P. TEDESCO, Indiana/Purdue University at Indianapolis CAROL MANKIEWICZ, Beloit College LORA SHRAKE, TA, Indiana/Purdue University at Indianapolis PROJECT OVERVIEW complicating environmental management are the needs of many federally and state-listed Southwest Florida encompasses one of the endangered species, including the Florida fastest growing regions in the United States. panther and West Indian manatee. Watershed The two southwestern coastal counties, Collier management must also consider these issues and Lee Counties, commonly make it among of environmental health and conservation. the 5 fastest growing population centers on nation- and statewide censuses. -

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Estero River for the Village of Estero
The Significance of the Estero River for the Village of Estero Photo: Florida Department of Environmental Protection Estuary = “Estero” Estero Bay Watershed Estero Bay Watershed: 360 square miles Pine Island Caloosahatchee Sound Matlacha Watershed Pass Watershed Estero River Basin: 66 square miles (45,000 acres) Estero River: Approximately 8 miles long Mean depth of 3.99 feet and a max depth of 12.06 feet (USF, 2013) Estero Bay Aquatic Preserve: 18,829 acres Source: Estero Bay Agency on Bay Management, State of the Bay Update 2014 Halfway Headwaters Wild Turkey Pond Strand Preserve Imperial Marsh Preserve To Estero River Flows (Source, South Lee County Watershed Plan Update, May 14, 2009) History Source: Estero Historical Society Estero River 1957, State Archives of Florida Historical Collection Estero River, Circa 1900 source: FGCU Collection / Koreshan “A Historical Geography of Southwest Florida Waterways , Vol. II Source: Estero Historical Society Pre-Development Conditions 1944 1953 1998 Courtesy: Lee County Natural Resources What happens in the DRGR affects Estero River and Estero Bay 1953 Hydropatterns 2007 Hydropatterns Source: Kevin Erwin, Consulting Ecologist Vegetation (Source: USF, West-Central Florida Tidal Stream Assessment Study, p. 123) Wildlife Listed Animal Species Gulf sturgeon roseate spoonbill American alligator peregrine falcon loggerhead turtle southeastern American kestrel common snook gopher tortoise Number of Known Atlantic green turtle American oystercatcher Native Species leatherback bald eagle Mammals: 37 -

Lascolabacillus Massiliensis'': a New Species Isolated
NEW MICROBES IN HUMANS “Lascolabacillus massiliensis”: a new consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 species isolated from the human gut (Marseille, France) were obtained under numbers 11-017 and 09-022, respectively. The initial growth was obtained after 10 days of culture in a 5% sheep blood-enriched sheep rumen M. Beye, S. Bakour, S. I. Traore, D. Raoult and P.-E. Fournier medium in aerobic atmosphere at 37°C. The bacterium was Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes, sub-cultured on 5% sheep blood-enriched Columbia agar Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille (bioMérieux, Marcy l’Etoile, France) and grew in 24 hours at Université, Faculté de Médecine, Marseille cedex 5, France 37°C in both aerobic and anaerobic conditions. Agar-grown colonies were pale grey and 1.5 mm in diameter. Bacterial cells were Gram-negative, rod-shaped and polymorphic, ranginginlengthfrom1.5to10μm. Strain SIT8 was catalase- Abstract and oxidase-negative. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, We report here the main characteristics of “Lascolabacillus France). Strain SIT8 exhibited a 94.14% sequence identity with massiliensis” strain SIT8 (CSUR P1560) that was isolated from the Proteiniphilum acetatigenes strain TB107 (GenBank accession stool of a healthy 28-month-old boy. NR_043154), the phylogenetically closest species with New Microbes and New Infections © 2016 The Authors. Published standing in nomenclature (Fig. 1), which putatively classifies it by Elsevier Ltd on behalf of European Society of Clinical as a member of a new genus within the family Porphyr- Microbiology and Infectious Diseases. -
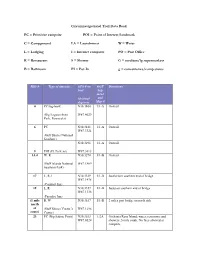
Circumnavigational Trail Data Book PC = Primitive Campsite POI
Circumnavigational Trail Data Book PC = Primitive campsite POI = Point of Interest/landmark C = Campground LA = Laundromat W = Water L = Lodging I = Internet computer PO = Post Office R = Restaurant S = Shower G = medium/lg supermarkets B = Bathroom PI = Put-In g = convenience/camp stores Mile # Type of amenity GPS # on OGT Directions trail Seg- ment (decimal- and degrees) Map # 0 PC/log-book N30.3100 1/1-A On trail (Big Lagoon State W87.4029 Park, Pensacola) 6 PC N30.3181 1/1-A On trail W87.3321 (Gulf Shores National Seashore) N30.3298 1/1-A On trail 8 POI (Ft. Pickens) W87.3018 14.4 W, R N30.3270 1/1-B On trail (Gulf Islands National W87.1809 Seashore Park) 17 L, R, I N30.3329 1/1-B Just before southern end of bridge W87.1438 (Comfort Inn) 18 L, R N30.3357 1/1-B Just past southern end of bridge W87.1338 (Paradise Inn) (1 mile R, W N30.3637 1/1-B 2 miles past bridge on north side north of (Gulf Shores Visitor’s W87.1296 route) Center) 23 PC (Big Sabine Point) N30.3533 1/2A On Santa Rosa Island; water, restrooms and W87.0524 showers .5 mile south. No fires allowed at campsite. (1 mile PI (no facilities) N30.3740 1/2A North side of sound north of W87.0924 route) 27.8 PI (no facilities) N30.3874 1/2A North side of sound W86.9967 35 G, L, I (Best Western), N30.4007 1/2B North side of sound just past bridge. -

Diversity and Composition of the Skin, Blood and Gut Microbiome in Rosacea—A Systematic Review of the Literature
microorganisms Review Diversity and Composition of the Skin, Blood and Gut Microbiome in Rosacea—A Systematic Review of the Literature Klaudia Tutka, Magdalena Zychowska˙ and Adam Reich * Department of Dermatology, Institute of Medical Sciences, Medical College of Rzeszow University, 35-055 Rzeszow, Poland; [email protected] (K.T.); [email protected] (M.Z.)˙ * Correspondence: [email protected]; Tel.: +48-605076722 Received: 30 August 2020; Accepted: 6 November 2020; Published: 8 November 2020 Abstract: Rosacea is a chronic inflammatory skin disorder of a not fully understood pathophysiology. Microbial factors, although not precisely characterized, are speculated to contribute to the development of the condition. The aim of the current review was to summarize the rosacea-associated alterations in the skin, blood, and gut microbiome, investigated using culture-independent, metagenomic techniques. A systematic review of the PubMed, Web of Science, and Scopus databases was performed, according to PRISMA (preferred reporting items for systematic review and meta-analyses) guidelines. Nine out of 185 papers were eligible for analysis. Skin microbiome was investigated in six studies, and in a total number of 115 rosacea patients. Blood microbiome was the subject of one piece of research, conducted in 10 patients with rosacea, and gut microbiome was studied in two papers, and in a total of 23 rosacea subjects. Although all of the studies showed significant alterations in the composition of the skin, blood, or gut microbiome in rosacea, the results were highly inconsistent, or even, in some cases, contradictory. Major limitations included the low number of participants, and different study populations (mainly Asians). Further studies are needed in order to reliably analyze the composition of microbiota in rosacea, and the potential application of microbiome modifications for the treatment of this dermatosis. -

State of the Bay Update Estero Bay Agency on Bay Manag on Bay Agency Bay Estero
ement 2009 State of the Bay Update Estero Bay Agency on Bay Manag on Bay Agency Bay Estero Southwest Florida Regional Planning Council 1926 Victoria Avenue, Fort Myers, Florida Contents Acknowledgements ................................................................................................................................................................. 4 Introduction ............................................................................................................................................................................ 5 Principles of the Estero Bay Agency on Bay Management ................................................................................................. 6 Human History of Estero Bay ................................................................................................................................................ 11 Water Quality ........................................................................................................................................................................ 19 2008 Water Quality Status ................................................................................................................................................ 19 Comparison of Water Quality Standards .......................................................................................................................... 20 Impaired Waters .............................................................................................................................................................. -

Friends of the Museum of the Everglades August 2017
Friends of the Museum of the Everglades August 2017 Volume XVII Issue 6 FME, P.O. Box 677, Everglades City, FL 34139 August 2017 Website: www.evergladesmuseum.org email: [email protected] Mark Your Calendar Pauline Reeves Gallery Board Meeting Everglades City Students’ Work Displayed September 5, 2017 During the summer months student artists from the Everglades City School had their work 3:30 pm City Hall displayed. Under the direction of their art teacher, Mr. Tribble, more than 30 students have their two dimension art work on display. From the abstract to realism, from portraits to Pauline Reeves Gallery landscapes the talent displayed by students from Kindergarten to grade 12 is inspiring. June & July One of the artists, Dylan Doster, whose work is on display received the Carol Moseman Student Art Work Scholarship Award in the amount of $500. The money will be sent to the Lorenzo Walker School in Naples where Dylan plans to further his studies. This scholarship is presented by the August Friends of the Museum each year to an aspiring artist. Gallery Closed for Renovation Volunteer Spotlight Georgia Garrett Inside This Issue Museuming p. 2 Koreshan Unity Franconian Museum Volunteer Spotlight p. 3 Bridging Gaps Director’s Note p. 4 A monument to the Chokoloskee Bridge is being used to bridge the museum’s new parking lot to the Museum of the Everglades. Built in 1955 the original bridge connected the small shell island of `` ` Chokoloskee to the mainland. This crosswalk will be finished soon making a wonderful addition to the museum. The Museum is located at 105 West Broadway, Everglades City, telephone (239) 695-0008 Open to the public Monday through Saturday, 9 am to 4 pm Volume XVII Issue 6 FME, P.O. -

Uncovering the Potential of Termite Gut Microbiome for Lignocellulose Bioconversion in Anaerobic Batch Bioreactors
ORIGINAL RESEARCH published: 22 December 2017 doi: 10.3389/fmicb.2017.02623 Uncovering the Potential of Termite Gut Microbiome for Lignocellulose Bioconversion in Anaerobic Batch Bioreactors Lucas Auer 1, Adèle Lazuka 1, David Sillam-Dussès 2, 3, Edouard Miambi 4, Michael O’Donohue 1 and Guillermina Hernandez-Raquet 1* 1 Laboratoire d’Ingénierie des Systèmes Biologiques et des Procédés, Université de Toulouse, Centre National de la Recherche Scientifique, Institut National de la Recherche Agronomique, INSA, Toulouse, France, 2 Laboratoire d’Éthologie Expérimentale et Comparée, Université Paris 13 - Sorbonne Paris Cité, Villetaneuse, France, 3 Institut d’Ecologie et des Sciences de l’Environnement de Paris, Institut de Recherche Pour le Développement – Sorbonne Universités, Bondy, France, 4 Institut d’Ecologie et des Sciences de l’Environnement de Paris, Université Paris-Est Créteil, Créteil, France Termites are xylophages, being able to digest a wide variety of lignocellulosic biomass including wood with high lignin content. This ability to feed on recalcitrant plant material Edited by: is the result of complex symbiotic relationships, which involve termite-specific gut Phil B. Pope, microbiomes. Therefore, these represent a potential source of microorganisms for the Norwegian University of Life Sciences, Norway bioconversion of lignocellulose in bioprocesses targeting the production of carboxylates. Reviewed by: In this study, gut microbiomes of four termite species were studied for their capacity to Karen Elizabeth Rossmassler, degrade wheat straw and produce carboxylates in controlled bioreactors. All of the gut Colorado State University, United States microbiomes successfully degraded lignocellulose and up to 45% w/w of wheat straw Zakee L. Sabree, degradation was observed, with the Nasutitermes ephratae gut-microbiome displaying The Ohio State University, the highest levels of wheat straw degradation, carboxylate production and enzymatic United States activity. -

Chapter 62-302: Surface Water Quality Standards
Presented below are water quality standards that are in effect for Clean Water Act purposes. EPA is posting these standards as a convenience to users and has made a reasonable effort to assure their accuracy. Additionally, EPA has made a reasonable effort to identify parts of the standards that are not approved, disapproved, or are otherwise not in effect for Clean Water Act purposes. blue – not a standard black – standard TCHAPTER 62-302: WATER QUALITY STANDARDS Effective April 30, 2018 In instances where the EPA has determined that certain provisions are not considered new or revised water quality standards, the Agency has attempted to indicate those in blue text. However, the font color indicated within this document, should not be interpreted as the official position of the Agency, and primarily addresses recent reviews by the EPA. For more detailed explanations on the EPA’s analysis and rationale related to decisions of new or revised water quality standards, see the Agency’s historical decision documents and associated records or contact the appropriate Agency staff. Additionally, arrow boxes found in the margins of this document direct the reader to other new or revised water quality standards which are related to provisions found in Chapter 62-302 but are found in documents outside of the regulations and are generally incorporated by reference. (See Attachments A-E of this document). The State of Florida recently revised the table of surface water criteria set out at section 62-302, F.A.C., to incorporate new human health criteria and designated uses promulgated by the state in 2016. -
Paddling Trails Leave No Trace Principles 5
This brochure made possible by: Florida Paddling Trails Leave No Trace Principles 5. Watch for motorboats. Stay to the right and turn the When you paddle, please observe these principles of Leave bow into their wake. Respect anglers. Paddle to the No Trace. For more information, log on to Leave No Trace shore opposite their lines. at www.lnt.org. 6. Respect wildlife. Do not approach or harass wildlife, as they can be dangerous. It’s illegal to feed them. q Plan Ahead and Prepare q Camp on Durable Surfaces 7. Bring a cell phone in case of an emergency. Cell q Dispose of Waste Properly phone coverage can be sporadic, so careful preparation q Leave What You Find and contingency plans should be made in lieu of relying on q Minimize Campfire Impacts cell phone reception. q Respect Wildlife FloridaPaddling Trails q Be Considerate of Other Visitors 8. If you are paddling on your own, give a reliable A Guide to Florida’s Top person your float plan before you leave and www.FloridaGreenwaysAndTrails.com leave a copy on the dash of your car. A float Canoeing & Kayaking Trails Trail Tips plan contains information about your trip in the event that When you paddle, please follow these tips. Water you do not return as scheduled. Don’t forget to contact the conditions vary and it will be up to you to be person you left the float plan with when you return. You can prepared for them. download a sample float plan at http://www.floridastateparks.org/wilderness/docs/FloatPlan.pdf. -

Floods in Florida Magnitude and Frequency
UNITED STATES EPARTMENT OF THE INTERIOR- ., / GEOLOGICAL SURVEY FLOODS IN FLORIDA MAGNITUDE AND FREQUENCY By R.W. Pride Prepared in cooperation with Florida State Road Department Open-file report 1958 MAR 2 CONTENTS Page Introduction. ........................................... 1 Acknowledgements ....................................... 1 Description of the area ..................................... 1 Topography ......................................... 2 Coastal Lowlands ..................................... 2 Central Highlands ..................................... 2 Tallahassee Hills ..................................... 2 Marianna Lowlands .................................... 2 Western Highlands. .................................... 3 Drainage basins ....................................... 3 St. Marys River. ......_.............................. 3 St. Johns River ...................................... 3 Lake Okeechobee and the everglades. ............................ 3 Peace River ....................................... 3 Withlacoochee River. ................................... 3 Suwannee River ...................................... 3 Ochlockonee River. .................................... 5 Apalachicola River .................................... 5 Choctawhatchee, Yellow, Blackwater, Escambia, and Perdido Rivers. ............. 5 Climate. .......................................... 5 Flood records ......................................... 6 Method of flood-frequency analysis ................................. 9 Flood frequency at a gaging