Neurobiology, 2009
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
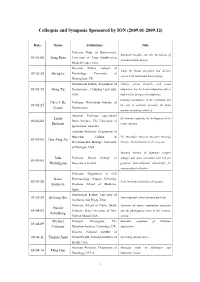
2009.01-2009.12)
Colloquia and Symposia Sponsored by ION (2009.01-2009.12) Date Name Affiliations Title Professor, Dept. of Biochemistry, Structural Insights into the Mechanism of 09-01-08 Josep Rizo University of Texas Southwestern Neurotransmitter Release. Medical Center, USA. Research Fellow, School of Study the human perception and decision 09-02-24 Sheng Li Psychology, University of systems with multimodal brain imaging. Birmingham, UK. Postdoctoral Fellow, Department of Probing cortical hierarchy with visual 09-02-24 Hong Xu Neuroscience, Columbia University, adaptation: low-level curve adaptation affects USA. high-level facial expression judgments. Creating coordination in the cerebellum and Chris I. De Professor, Netherlands Institute of 09-02-25 the role of artificial networks for brain Zeeuw Neuroscience. machine interfacing (robotics). Associate Professor, Queensland Linda Mechanisms regulating the development of the 09-03-02 Brain Institute, The University of Richards corpus callosum. Queensland, Australia. Assistant Professor, Department of Molecular, Cellular & The Mucolipin Transient Receptor Potential 09-03-02 Hao-Xing Xu Developmental Biology, University Proteins, I(r)on Channels in the Lysosome. of Michigan, USA. Targeted deletion of dopamine receptor John Professor, Royal College of subtypes and genes associated with risk for 09-03-05 Waddington Surgeons in Ireland. psychosis: brain-behaviour relationships for neuropsychiatric disorders. Professor, Department of Cell Kozo Pharmacology, Nagoya University 09-03-20 Axon formation and polarized transport. Kaibuchi Graduate School of Medicine, Japan. Postdoctoral Fellow, University of 09-03-24 Zi-Long Qiu Transcriptional control of neural plasticity. California, San Diego, USA. Professor, School of Public Health Functions of mature mammalian astrocytes Harold 09-04-03 Sciences, State University of New and the philosophical limits of the scientific Kimelberg York at Albany, USA. -

The Journal of Neuroscience May 30, 2007 • Volume 27 Number 22
The Journal of Neuroscience May 30, 2007 • Volume 27 Number 22 www.jneurosci.org i This Week in The Journal Toolbox 5837 Antigen-Retrieval Procedure for Bromodeoxyuridine Immunolabeling with Concurrent Labeling of Nuclear DNA and Antigens Damaged by HCl Pretreatment Xiaobing Tang, Douglas L. Falls, Xuekun Li, Tracy Lane, and Marla B. Luskin Journal Club 5845 A Critical Time for New Neurons in the Adult Hippocampus Benedetta Leuner, Erica R. Glasper, and Christian Mirescu Cover legend: Rapid retraction of presynaptic motor 5847 The Role of the Mirror System in Mapping Complex Sounds into Actions axon terminals after postsynaptic protein synthesis Alessandro D’Ausilio inhibition. A cell-impermeant protein synthesis inhibitor is injected along with a fluorescent dextran Brief Communications near the neuromuscular junction of a muscle fiber in a 5879 Multisensory Integration Shortens Physiological Response Latencies living adult mouse. Three days later, the motor axon Benjamin A. Rowland, Stephan Quessy, Terrence R. Stanford, and Barry E. Stein (yellow–green) innervating the injected muscle fiber (magenta) has become thin and has retracted from 5976 Blood Vessels Form a Scaffold for Neuroblast Migration in the Adult Olfactory Bulb postsynaptic acetylcholine receptors (red), whereas Serena Bovetti, Yi-Chun Hsieh, Patrizia Bovolin, Isabelle Perroteau, surrounding motor axons (cyan) show normal caliber Toida Kazunori, and Adam C. Puche and full occupation of receptor sites. For more information, see the article by McCann et al. in this 5981 Dissociating Task Performance from fMRI Repetition Attenuation in Ventral Visual issue (pages 6064–6067). Cortex Yaoda Xu, Nicholas B. Turk-Browne, and Marvin M. Chun 6064 Rapid Synapse Elimination After Postsynaptic Protein Synthesis Inhibition In Vivo Corey M. -

Spatiotemporal Dynamics of Dendritic Spines in the Living Brain
MINI REVIEW ARTICLE published: 09 May 2014 doi: 10.3389/fnana.2014.00028 Spatiotemporal dynamics of dendritic spines in the living brain Chia-Chien Chen1,JuLu2 andYi Zuo 1* 1 Department of Molecular, Cell and Developmental Biology, University of California at Santa Cruz, Santa Cruz, CA, USA 2 Department of Biological Sciences and James H. Clark Center, Stanford University, Stanford, CA, USA Edited by: Dendritic spines are ubiquitous postsynaptic sites of most excitatory synapses in the Nicolas Heck, University Pierre and mammalian brain, and thus may serve as structural indicators of functional synapses. Marie Curie, France Recent works have suggested that neuronal coding of memories may be associated Reviewed by: with rapid alterations in spine formation and elimination. Technological advances have Ricardo Insausti, University of Castilla-La Mancha, Spain enabled researchers to study spine dynamics in vivo during development as well as under Anthony Holtmaat, Department of various physiological and pathological conditions. We believe that better understanding Basic Neurosciences, Switzerland of the spatiotemporal patterns of spine dynamics will help elucidate the principles of *Correspondence: experience-dependent circuit modification and information processing in the living brain. Yi Zuo, Department of Molecular, Cell and Developmental Biology, Keywords: dendritic spine, in vivo, two-photon imaging, experience-dependent plasticity, neural circuit, cerebral University of California at Santa Cruz, cortex 1156 High Street, Santa Cruz, CA 95064, USA e-mail: [email protected] INTRODUCTION experiences such as sensory stimulation and deprivation, environ- Dendritic spines have fascinated generations of neuroscientists mental enrichment, and various paradigms of learning (Holtmaat since their initial description by Santiago Ramón y Cajal more and Svoboda, 2009; Fu and Zuo, 2011). -

Multifaceted Changes in Synaptic Composition and Astrocytic Involvement in a Mouse Model of Fragile X Syndrome
UC Santa Cruz UC Santa Cruz Previously Published Works Title Multifaceted Changes in Synaptic Composition and Astrocytic Involvement in a Mouse Model of Fragile X Syndrome. Permalink https://escholarship.org/uc/item/79v7g3g9 Journal Scientific reports, 9(1) ISSN 2045-2322 Authors Simhal, Anish K Zuo, Yi Perez, Marc M et al. Publication Date 2019-09-25 DOI 10.1038/s41598-019-50240-x Peer reviewed eScholarship.org Powered by the California Digital Library University of California www.nature.com/scientificreports OPEN Multifaceted Changes in Synaptic Composition and Astrocytic Involvement in a Mouse Model of Received: 14 May 2019 Accepted: 9 September 2019 Fragile X Syndrome Published: xx xx xxxx Anish K. Simhal 1, Yi Zuo 2, Marc M. Perez3, Daniel V. Madison3, Guillermo Sapiro1,4 & Kristina D. Micheva3 Fragile X Syndrome (FXS), a common inheritable form of intellectual disability, is known to alter neocortical circuits. However, its impact on the diverse synapse types comprising these circuits, or on the involvement of astrocytes, is not well known. We used immunofuorescent array tomography to quantify diferent synaptic populations and their association with astrocytes in layers 1 through 4 of the adult somatosensory cortex of a FXS mouse model, the FMR1 knockout mouse. The collected multi- channel data contained approximately 1.6 million synapses which were analyzed using a probabilistic synapse detector. Our study reveals complex, synapse-type and layer specifc changes in the neocortical circuitry of FMR1 knockout mice. We report an increase of small glutamatergic VGluT1 synapses in layer 4 accompanied by a decrease in large VGluT1 synapses in layers 1 and 4. -

The Johns Hopkins University
THE JOHNS HOPKINS UNIVERSITY COMMENCEMENT 2019 Conferring of degrees at the close of the 143rd academic year MAY 23, 2019 CONTENTS Order of Candidate Procession ......................................................... 11 Order of Procession .......................................................................... 12 Order of Events ................................................................................. 13 Conferring of Degrees ....................................................................... 14 Honorary Degrees ............................................................................. 17 Academic Garb .................................................................................. 14 Awards ............................................................................................... 16 Honor Societies ................................................................................. 28 Student Honors ................................................................................. 34 Candidates for Degrees ..................................................................... 43 Divisional Ceremonies Information ............................................... 121 Please note that while all degrees are conferred, only doctoral and bachelor’s graduates process across the stage. Taking photos from your seats during the ceremony is allowed, but we request that guests respect one another’s comfort and enjoyment by not standing and blocking other people’s views. Photos of graduates can be purchased from GradImages®: gradimages.com -

Uncorrected Proof
Biochemical Pharmacology xxx (xxxx) xxx-xxx Contents lists available at ScienceDirect Biochemical Pharmacology journal homepage: http://ees.elsevier.com Review Post-learning micro- and macro-structural neuroplasticity changes with time and sleep Whitney Stee a,b,⁎, Philippe Peigneux a,b,⁎ a UR2NF-Neuropsychology and Functional Neuroimaging Research Unit Affiliated at CRCN – Centre for Research in Cognition and Neurosciences, Avenue F.D, Roosevelt 50, Bruxelles 1050, Belgium b UNI-ULB Neuroscience Institute, Université Libre de Bruxelles (ULB), Avenue F.D. Roosevelt 50, Bruxelles 1050, Belgium ARTICLE INFO ABSTRACT PROOF Keywords Neuroplasticity refers to the fact that our brain can partially modify both structure and function to adequately Structural neuroimaging respond to novel environmental stimulations. Neuroplasticity mechanisms are not only operating during the ac- Diffusion weighted imaging (DWI) quisition of novel information (i.e., online) but also during the offline periods that take place after the end of the Neuroplasticity actual learning episode. Structural brain changes as a consequence of learning have been consistently demon- Learning and memory consolidation strated on the long term using non-invasive neuroimaging methods, but short-term changes remained more elu- Sleep sive. Fortunately, the swift development of advanced MR methods over the last decade now allows tracking fine-grained cerebral changes on short timescales beyond gross volumetric modifications stretching over several days or weeks. Besides a mere effect of time, post-learning sleep mechanisms have been shown to play an im- portant role in memory consolidation and promote long-lasting changes in neural networks. Sleep was shown to contribute to structural modifications over weeks of prolonged training, but studies evidencing more rapid post-training sleep structural effects linked to memory consolidation are still scarce in human. -

Page 1 of 12 CURRICULUM VITAE Theresa A. Jones January 2021 Psychology Department University of Texas Education Ph.D. 1992 Un
CURRICULUM VITAE Theresa A. Jones January 2021 Psychology Department University of Texas Education Ph.D. 1992 University of Texas at Austin, Behavioral Neuroscience Area of Psychology B.A. 1987 University of Texas at Austin, Psychology 1983-1986 University of Missouri-Columbia, Psychology Professional Experience 2001- Assistant (2001-03), Associate (2003-08), Full (2008) Professor, Behavioral Neuroscience Area Head (2009-13), Psychology Department and Institute for Neuroscience, The University of Texas at Austin 2013-2014 Bergeron Visiting Professor, Center for Brain Plasticity and Behavior, Georgetown University Medical Center, Washington, D.C. 1996-2001 Assistant Professor, Psychology Department and Neurobiology and Behavior Program, University of Washington 1993-1996 Postdoctoral Fellow, Behavioral Neuroscience and Biopsychology, Beckman Institute, University of Illinois, Mentor: William T. Greenough 1987-1992 Graduate Student, Psychology Department and Neuroscience Institute, University of Texas at Austin, Mentor: Timothy Schallert Researcher Profiles Google Scholar researcher profile: https://scholar.google.com/citations?user=9hQ2fv8AAAAJ&hl=en Publons/Web of Science ResearcherID: https://publons.com/researcher/2831601/theresa-a-jones/ ORCID ID: https://orcid.org/0000-0003-0906-6439 AWARDS Research Funding Current NINDS R37 NS056839 (Javits Award, Formerly RO1 NS056839 & RO1 MH/NS064586) Neural mechanisms of compensating for brain damage, PI, 2002-26 NINDS R21 NS101564 Sex-dependent aging effects on cortical reorganization after -

Basal and Experience Dependent Ampar and Synapse Dynamics: Alterations in a Mouse Model of Fragile X Syndrome
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 Basal and Experience Dependent Ampar and Synapse Dynamics: Alterations in a Mouse Model of Fragile X Syndrome Anand Suresh University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Molecular and Cellular Neuroscience Commons Recommended Citation Suresh, Anand, "Basal and Experience Dependent Ampar and Synapse Dynamics: Alterations in a Mouse Model of Fragile X Syndrome" (2017). Theses & Dissertations. 179. https://digitalcommons.unmc.edu/etd/179 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. i BASAL AND EXPERIENCE DEPENDENT AMPAR AND SYNAPSE DYNAMICS: ALTERATIONS IN A MOUSE MODEL OF FRAGILE X SYNDROME By Anand Suresh A Dissertation Presented to the Faculty of The Graduate College in the University of Nebraska Medical Center In partial Fulfillment of the Requirements For the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Under the supervision of Professor Anna Dunaevsky University of Nebraska Medical Center Omaha, Nebraska December 2016 ii Acknowledgements I would like to first thank my mentor, Dr. Anna Dunaevsky for her patience, support and guidance throughout my graduate studies. Her immense drive and passion towards biomedical research has been a constant source of inspiration for me and will remain so for the rest of my scientific career. I am also grateful for her endless investment in mine and other trainee’s professional advancement. -

2014-15 General Catalog Faculty Chancellor's Welcome
Faculty University Home MyUCSC People Calendars A-Z Index OFFICE OF THE REGISTRAR ABOUT CALENDAR FEES ENROLLMENT RECORDS QUICK START GUIDE Home » UCSC General Catalog » Programs and Courses » Faculty 2014-15 General Catalog Faculty Chancellor's Welcome About For faculty listings, see the program statement for individual programs and departments. Fields of Study Calendar Undergraduate Admission Undergraduate Expenses Undergraduate Academic Program Graduate Studies Resources for Learning and Research Campus Life Programs and Courses Disciplinary Communication General Education Requirements by Department Program Statements Faculty Anthropology Applied Mathematics and Statistics Art Astronomy and Astrophysics Bioengineering Biochemistry and Molecular Biology Biomolecular Engineering Chemistry and Biochemistry Chinese Classical Studies Community Studies Computer Engineering Computer Science Critical Race and Ethnic Studies Digital Arts and New Media Earth and Planetary Science East Asian Studies Economics http://registrar.ucsc.edu/catalog/programs-courses/faculty/index.html[8/14/2014 10:35:13 AM] Anthropology University Home MyUCSC People Calendars A-Z Index OFFICE OF THE REGISTRAR ABOUT CALENDAR FEES ENROLLMENT RECORDS QUICK START GUIDE Home » UCSC General Catalog » Programs and Courses » Faculty » Anthropology 2014-15 General Catalog Anthropology Chancellor's Welcome About 2014-15 General Catalog Fields of Study 361 Social Sciences 1 Building Calendar (831) 459- 3320 http://anthro.ucsc.edu/ Undergraduate Admission Program Description | Course -

Pyramidal Neurons in Different Cortical Layers Exhibit Distinct Dynamics and Plasticity of Apical Dendritic Spines
UC Santa Cruz UC Santa Cruz Previously Published Works Title Pyramidal Neurons in Different Cortical Layers Exhibit Distinct Dynamics and Plasticity of Apical Dendritic Spines. Permalink https://escholarship.org/uc/item/4zr002gw Authors Tjia, Michelle Yu, Xinzhu Jammu, Lavpreet S et al. Publication Date 2017 DOI 10.3389/fncir.2017.00043 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ORIGINAL RESEARCH published: 19 June 2017 doi: 10.3389/fncir.2017.00043 Pyramidal Neurons in Different Cortical Layers Exhibit Distinct Dynamics and Plasticity of Apical Dendritic Spines Michelle Tjia, Xinzhu Yu, Lavpreet S. Jammu, Ju Lu and Yi Zuo* Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, CA, United States The mammalian cerebral cortex is typically organized in six layers containing multiple types of neurons, with pyramidal neurons (PNs) being the most abundant. PNs in different cortical layers have distinct morphology, physiology and functional roles in neural circuits. Therefore, their development and synaptic plasticity may also differ. Using in vivo transcranial two-photon microscopy, we followed the structural dynamics of dendritic spines on apical dendrites of layer (L) 2/3 and L5 PNs at different developmental stages. We show that the density and dynamics of spines are significantly higher in L2/3 PNs than L5 PNs in both adolescent (1 month old) and adult (4 months old) mice. While spine density of L5 PNs decreases during adolescent development due to a higher rate of spine elimination than formation, there is no net change in the spine density along apical dendrites of L2/3 PNs over this period. -

An Analog of Psychedelics Restores Functional Neural Circuits Disrupted by Unpredictable Stress
Molecular Psychiatry https://doi.org/10.1038/s41380-021-01159-1 ARTICLE An analog of psychedelics restores functional neural circuits disrupted by unpredictable stress 1 1 1 2 1 1 Ju Lu ● Michelle Tjia ● Brian Mullen ● Bing Cao ● Kacper Lukasiewicz ● Sajita Shah-Morales ● 1 3 4,5,6 2 1 Sydney Weiser ● Lindsay P. Cameron ● David E. Olson ● Lu Chen ● Yi Zuo Received: 7 January 2021 / Revised: 28 April 2021 / Accepted: 5 May 2021 © The Author(s) 2021. This article is published with open access Abstract Psychological stress affects a wide spectrum of brain functions and poses risks for many mental disorders. However, effective therapeutics to alleviate or revert its deleterious effects are lacking. A recently synthesized psychedelic analog tabernanthalog (TBG) has demonstrated anti-addictive and antidepressant potential. Whether TBG can rescue stress-induced affective, sensory, and cognitive deficits, and how it may achieve such effects by modulating neural circuits, remain unknown. Here we show that in mice exposed to unpredictable mild stress (UMS), administration of a single dose of TBG decreases their anxiety level and rescues deficits in sensory processing as well as in cognitive flexibility. Post-stress TBG 1234567890();,: 1234567890();,: treatment promotes the regrowth of excitatory neuron dendritic spines lost during UMS, decreases the baseline neuronal activity, and enhances whisking-modulation of neuronal activity in the somatosensory cortex. Moreover, calcium imaging in head-fixed mice performing a whisker-dependent texture discrimination task shows that novel textures elicit responses from a greater proportion of neurons in the somatosensory cortex than do familiar textures. Such differential response is diminished by UMS and is restored by TBG. -

Spatial Constraints Dictate Glial Territories at Murine Neuromuscular Junctions
Spatial Constraints Dictate Glial Territories at Murine Neuromuscular Junctions The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Brill, Monika S., Jeff W. Lichtman, Wesley Thompson, Yi Zuo, and Thomas Misgeld. 2011. Spatial constraints dictate glial territories at murine neuromuscular junctions. The Journal of Cell Biology 195(2): 293-305. Published Version doi:10.1083/jcb.201108005 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:11210599 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA JCB: Article Spatial constraints dictate glial territories at murine neuromuscular junctions Monika S. Brill,1 Jeff W. Lichtman,2 Wesley Thompson,3 Yi Zuo,4 and Thomas Misgeld1,5 1Chair of Biomolecular Sensors, Center for Integrated Protein Science Munich at the Institute of Neuroscience, Technische Universität München, 80802 Munich, Germany 2Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138 3Institute for Neuroscience, University of Texas at Austin, Austin, TX 78712 4Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, Santa Cruz, CA 95064 5Institute for Advanced Study, Technische Universität München, 85748 Garching bei München, Germany chwann cells (SCs), the glial cells of the peripheral in which glial–glial and axonal–glial contacts constrain nervous system, cover synaptic terminals, allowing the territory of single SCs, as shown by four types of S them to monitor and modulate neurotransmission.