Python for Data Science Cheat Sheet
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sagemath and Sagemathcloud
Viviane Pons Ma^ıtrede conf´erence,Universit´eParis-Sud Orsay [email protected] { @PyViv SageMath and SageMathCloud Introduction SageMath SageMath is a free open source mathematics software I Created in 2005 by William Stein. I http://www.sagemath.org/ I Mission: Creating a viable free open source alternative to Magma, Maple, Mathematica and Matlab. Viviane Pons (U-PSud) SageMath and SageMathCloud October 19, 2016 2 / 7 SageMath Source and language I the main language of Sage is python (but there are many other source languages: cython, C, C++, fortran) I the source is distributed under the GPL licence. Viviane Pons (U-PSud) SageMath and SageMathCloud October 19, 2016 3 / 7 SageMath Sage and libraries One of the original purpose of Sage was to put together the many existent open source mathematics software programs: Atlas, GAP, GMP, Linbox, Maxima, MPFR, PARI/GP, NetworkX, NTL, Numpy/Scipy, Singular, Symmetrica,... Sage is all-inclusive: it installs all those libraries and gives you a common python-based interface to work on them. On top of it is the python / cython Sage library it-self. Viviane Pons (U-PSud) SageMath and SageMathCloud October 19, 2016 4 / 7 SageMath Sage and libraries I You can use a library explicitly: sage: n = gap(20062006) sage: type(n) <c l a s s 'sage. interfaces .gap.GapElement'> sage: n.Factors() [ 2, 17, 59, 73, 137 ] I But also, many of Sage computation are done through those libraries without necessarily telling you: sage: G = PermutationGroup([[(1,2,3),(4,5)],[(3,4)]]) sage : G . g a p () Group( [ (3,4), (1,2,3)(4,5) ] ) Viviane Pons (U-PSud) SageMath and SageMathCloud October 19, 2016 5 / 7 SageMath Development model Development model I Sage is developed by researchers for researchers: the original philosophy is to develop what you need for your research and share it with the community. -

A Comparative Evaluation of Matlab, Octave, R, and Julia on Maya 1 Introduction
A Comparative Evaluation of Matlab, Octave, R, and Julia on Maya Sai K. Popuri and Matthias K. Gobbert* Department of Mathematics and Statistics, University of Maryland, Baltimore County *Corresponding author: [email protected], www.umbc.edu/~gobbert Technical Report HPCF{2017{3, hpcf.umbc.edu > Publications Abstract Matlab is the most popular commercial package for numerical computations in mathematics, statistics, the sciences, engineering, and other fields. Octave is a freely available software used for numerical computing. R is a popular open source freely available software often used for statistical analysis and computing. Julia is a recent open source freely available high-level programming language with a sophisticated com- piler for high-performance numerical and statistical computing. They are all available to download on the Linux, Windows, and Mac OS X operating systems. We investigate whether the three freely available software are viable alternatives to Matlab for uses in research and teaching. We compare the results on part of the equipment of the cluster maya in the UMBC High Performance Computing Facility. The equipment has 72 nodes, each with two Intel E5-2650v2 Ivy Bridge (2.6 GHz, 20 MB cache) proces- sors with 8 cores per CPU, for a total of 16 cores per node. All nodes have 64 GB of main memory and are connected by a quad-data rate InfiniBand interconnect. The tests focused on usability lead us to conclude that Octave is the most compatible with Matlab, since it uses the same syntax and has the native capability of running m-files. R was hampered by somewhat different syntax or function names and some missing functions. -

Data Visualization in Python
Data visualization in python Day 2 A variety of packages and philosophies • (today) matplotlib: http://matplotlib.org/ – Gallery: http://matplotlib.org/gallery.html – Frequently used commands: http://matplotlib.org/api/pyplot_summary.html • Seaborn: http://stanford.edu/~mwaskom/software/seaborn/ • ggplot: – R version: http://docs.ggplot2.org/current/ – Python port: http://ggplot.yhathq.com/ • Bokeh (live plots in your browser) – http://bokeh.pydata.org/en/latest/ Biocomputing Bootcamp 2017 Matplotlib • Gallery: http://matplotlib.org/gallery.html • Top commands: http://matplotlib.org/api/pyplot_summary.html • Provides "pylab" API, a mimic of matlab • Many different graph types and options, some obscure Biocomputing Bootcamp 2017 Matplotlib • Resulting plots represented by python objects, from entire figure down to individual points/lines. • Large API allows any aspect to be tweaked • Lengthy coding sometimes required to make a plot "just so" Biocomputing Bootcamp 2017 Seaborn • https://stanford.edu/~mwaskom/software/seaborn/ • Implements more complex plot types – Joint points, clustergrams, fitted linear models • Uses matplotlib "under the hood" Biocomputing Bootcamp 2017 Others • ggplot: – (Original) R version: http://docs.ggplot2.org/current/ – A recent python port: http://ggplot.yhathq.com/ – Elegant syntax for compactly specifying plots – but, they can be hard to tweak – We'll discuss this on the R side tomorrow, both the basics of both work similarly. • Bokeh – Live, clickable plots in your browser! – http://bokeh.pydata.org/en/latest/ -

Mastering Machine Learning with Scikit-Learn
www.it-ebooks.info Mastering Machine Learning with scikit-learn Apply effective learning algorithms to real-world problems using scikit-learn Gavin Hackeling BIRMINGHAM - MUMBAI www.it-ebooks.info Mastering Machine Learning with scikit-learn Copyright © 2014 Packt Publishing All rights reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, without the prior written permission of the publisher, except in the case of brief quotations embedded in critical articles or reviews. Every effort has been made in the preparation of this book to ensure the accuracy of the information presented. However, the information contained in this book is sold without warranty, either express or implied. Neither the author, nor Packt Publishing, and its dealers and distributors will be held liable for any damages caused or alleged to be caused directly or indirectly by this book. Packt Publishing has endeavored to provide trademark information about all of the companies and products mentioned in this book by the appropriate use of capitals. However, Packt Publishing cannot guarantee the accuracy of this information. First published: October 2014 Production reference: 1221014 Published by Packt Publishing Ltd. Livery Place 35 Livery Street Birmingham B3 2PB, UK. ISBN 978-1-78398-836-5 www.packtpub.com Cover image by Amy-Lee Winfield [email protected]( ) www.it-ebooks.info Credits Author Project Coordinator Gavin Hackeling Danuta Jones Reviewers Proofreaders Fahad Arshad Simran Bhogal Sarah -

Installing Python Ø Basics of Programming • Data Types • Functions • List Ø Numpy Library Ø Pandas Library What Is Python?
Python in Data Science Programming Ha Khanh Nguyen Agenda Ø What is Python? Ø The Python Ecosystem • Installing Python Ø Basics of Programming • Data Types • Functions • List Ø NumPy Library Ø Pandas Library What is Python? • “Python is an interpreted high-level general-purpose programming language.” – Wikipedia • It supports multiple programming paradigms, including: • Structured/procedural • Object-oriented • Functional The Python Ecosystem Source: Fabien Maussion’s “Getting started with Python” Workshop Installing Python • Install Python through Anaconda/Miniconda. • This allows you to create and work in Python environments. • You can create multiple environments as needed. • Highly recommended: install Python through Miniconda. • Are we ready to “play” with Python yet? • Almost! • Most data scientists use Python through Jupyter Notebook, a web application that allows you to create a virtual notebook containing both text and code! • Python Installation tutorial: [Mac OS X] [Windows] For today’s seminar • Due to the time limitation, we will be using Google Colab instead. • Google Colab is a free Jupyter notebook environment that runs entirely in the cloud, so you can run Python code, write report in Jupyter Notebook even without installing the Python ecosystem. • It is NOT a complete alternative to installing Python on your local computer. • But it is a quick and easy way to get started/try out Python. • You will need to log in using your university account (possible for some schools) or your personal Google account. Basics of Programming Indentations – Not Braces • Other languages (R, C++, Java, etc.) use braces to structure code. # R code (not Python) a = 10 if (a < 5) { result = TRUE b = 0 } else { result = FALSE b = 100 } a = 10 if (a < 5) { result = TRUE; b = 0} else {result = FALSE; b = 100} Basics of Programming Indentations – Not Braces • Python uses whitespaces (tabs or spaces) to structure code instead. -

How to Access Python for Doing Scientific Computing
How to access Python for doing scientific computing1 Hans Petter Langtangen1,2 1Center for Biomedical Computing, Simula Research Laboratory 2Department of Informatics, University of Oslo Mar 23, 2015 A comprehensive eco system for scientific computing with Python used to be quite a challenge to install on a computer, especially for newcomers. This problem is more or less solved today. There are several options for getting easy access to Python and the most important packages for scientific computations, so the biggest issue for a newcomer is to make a proper choice. An overview of the possibilities together with my own recommendations appears next. Contents 1 Required software2 2 Installing software on your laptop: Mac OS X and Windows3 3 Anaconda and Spyder4 3.1 Spyder on Mac............................4 3.2 Installation of additional packages.................5 3.3 Installing SciTools on Mac......................5 3.4 Installing SciTools on Windows...................5 4 VMWare Fusion virtual machine5 4.1 Installing Ubuntu...........................6 4.2 Installing software on Ubuntu....................7 4.3 File sharing..............................7 5 Dual boot on Windows8 6 Vagrant virtual machine9 1The material in this document is taken from a chapter in the book A Primer on Scientific Programming with Python, 4th edition, by the same author, published by Springer, 2014. 7 How to write and run a Python program9 7.1 The need for a text editor......................9 7.2 Spyder................................. 10 7.3 Text editors.............................. 10 7.4 Terminal windows.......................... 11 7.5 Using a plain text editor and a terminal window......... 12 8 The SageMathCloud and Wakari web services 12 8.1 Basic intro to SageMathCloud................... -

An Introduction to Numpy and Scipy
An introduction to Numpy and Scipy Table of contents Table of contents ............................................................................................................................ 1 Overview ......................................................................................................................................... 2 Installation ...................................................................................................................................... 2 Other resources .............................................................................................................................. 2 Importing the NumPy module ........................................................................................................ 2 Arrays .............................................................................................................................................. 3 Other ways to create arrays............................................................................................................ 7 Array mathematics .......................................................................................................................... 8 Array iteration ............................................................................................................................... 10 Basic array operations .................................................................................................................. 11 Comparison operators and value testing .................................................................................... -
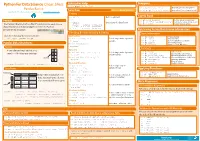
Cheat Sheet Pandas Python.Indd
Python For Data Science Cheat Sheet Asking For Help Dropping >>> help(pd.Series.loc) Pandas Basics >>> s.drop(['a', 'c']) Drop values from rows (axis=0) Selection Also see NumPy Arrays >>> df.drop('Country', axis=1) Drop values from columns(axis=1) Learn Python for Data Science Interactively at www.DataCamp.com Getting Sort & Rank >>> s['b'] Get one element -5 Pandas >>> df.sort_index() Sort by labels along an axis Get subset of a DataFrame >>> df.sort_values(by='Country') Sort by the values along an axis The Pandas library is built on NumPy and provides easy-to-use >>> df[1:] Assign ranks to entries Country Capital Population >>> df.rank() data structures and data analysis tools for the Python 1 India New Delhi 1303171035 programming language. 2 Brazil Brasília 207847528 Retrieving Series/DataFrame Information Selecting, Boolean Indexing & Setting Basic Information Use the following import convention: By Position (rows,columns) Select single value by row & >>> df.shape >>> import pandas as pd >>> df.iloc[[0],[0]] >>> df.index Describe index Describe DataFrame columns 'Belgium' column >>> df.columns Pandas Data Structures >>> df.info() Info on DataFrame >>> df.iat([0],[0]) Number of non-NA values >>> df.count() Series 'Belgium' Summary A one-dimensional labeled array a 3 By Label Select single value by row & >>> df.sum() Sum of values capable of holding any data type b -5 >>> df.loc[[0], ['Country']] Cummulative sum of values 'Belgium' column labels >>> df.cumsum() >>> df.min()/df.max() Minimum/maximum values c 7 Minimum/Maximum index -

Homework 2 an Introduction to Convolutional Neural Networks
Homework 2 An Introduction to Convolutional Neural Networks 11-785: Introduction to Deep Learning (Spring 2019) OUT: February 17, 2019 DUE: March 10, 2019, 11:59 PM Start Here • Collaboration policy: { You are expected to comply with the University Policy on Academic Integrity and Plagiarism. { You are allowed to talk with / work with other students on homework assignments { You can share ideas but not code, you must submit your own code. All submitted code will be compared against all code submitted this semester and in previous semesters using MOSS. • Overview: { Part 1: All of the problems in Part 1 will be graded on Autolab. You can download the starter code from Autolab as well. This assignment has 100 points total. { Part 2: This section of the homework is an open ended competition hosted on Kaggle.com, a popular service for hosting predictive modeling and data analytic competitions. All of the details for completing Part 2 can be found on the competition page. • Submission: { Part 1: The compressed handout folder hosted on Autolab contains four python files, layers.py, cnn.py, mlp.py and local grader.py, and some folders data, autograde and weights. File layers.py contains common layers in convolutional neural networks. File cnn.py contains outline class of two convolutional neural networks. File mlp.py contains a basic MLP network. File local grader.py is foryou to evaluate your own code before submitting online. Folders contain some data to be used for section 1.3 and 1.4 of this homework. Make sure that all class and function definitions originally specified (as well as class attributes and methods) in the two files layers.py and cnn.py are fully implemented to the specification provided in this write-up. -

Principal Components Analysis Tutorial 4 Yang
Principal Components Analysis Tutorial 4 Yang 1 Objectives Understand the principles of principal components analysis (PCA) Know the principal components analysis method Study the PCA function of sickit-learn.decomposition Process the data set by the PCA of sickit-learn Learn to apply PCA in a reality example 2 Principal Components Analysis Method: ̶ Subtract the mean ̶ Calculate the covariance matrix ̶ Calculate the eigenvectors and eigenvalues of the covariance matrix ̶ Choosing components and forming a feature vector ̶ Deriving the new data set 3 Example 1 x y 퐷푎푡푎 = 2.5 2.4 0.5 0.7 2.2 2.9 1.9 2.2 3.1 3.0 2.3 2.7 2 1.6 1 1.1 1.5 1.6 1.1 0.9 4 sklearn.decomposition.PCA It uses the LAPACK implementation of the full SVD or a randomized truncated SVD by the method of Halko et al. 2009, depending on the shape of the input data and the number of components to extract. It can also use the scipy.sparse.linalg ARPACK implementation of the truncated SVD. Notice that this class does not upport sparse input. 5 Parameters of PCA n_components: Number of components to keep. svd_solver: if n_components is not set: n_components == auto: default, if the input data is larger than 500x500 min (n_samples, n_features), default=None and the number of components to extract is lower than 80% of the smallest dimension of the data, then the more efficient ‘randomized’ method is enabled. if n_components == ‘mle’ and svd_solver == ‘full’, Otherwise the exact full SVD is computed and Minka’s MLE is used to guess the dimension optionally truncated afterwards. -

User Guide Release 1.18.4
NumPy User Guide Release 1.18.4 Written by the NumPy community May 24, 2020 CONTENTS 1 Setting up 3 2 Quickstart tutorial 9 3 NumPy basics 33 4 Miscellaneous 97 5 NumPy for Matlab users 103 6 Building from source 111 7 Using NumPy C-API 115 Python Module Index 163 Index 165 i ii NumPy User Guide, Release 1.18.4 This guide is intended as an introductory overview of NumPy and explains how to install and make use of the most important features of NumPy. For detailed reference documentation of the functions and classes contained in the package, see the reference. CONTENTS 1 NumPy User Guide, Release 1.18.4 2 CONTENTS CHAPTER ONE SETTING UP 1.1 What is NumPy? NumPy is the fundamental package for scientific computing in Python. It is a Python library that provides a multidi- mensional array object, various derived objects (such as masked arrays and matrices), and an assortment of routines for fast operations on arrays, including mathematical, logical, shape manipulation, sorting, selecting, I/O, discrete Fourier transforms, basic linear algebra, basic statistical operations, random simulation and much more. At the core of the NumPy package, is the ndarray object. This encapsulates n-dimensional arrays of homogeneous data types, with many operations being performed in compiled code for performance. There are several important differences between NumPy arrays and the standard Python sequences: • NumPy arrays have a fixed size at creation, unlike Python lists (which can grow dynamically). Changing the size of an ndarray will create a new array and delete the original. -

Introduction to Python in HPC
Introduction to Python in HPC Omar Padron National Center for Supercomputing Applications University of Illinois at Urbana Champaign [email protected] Outline • What is Python? • Why Python for HPC? • Python on Blue Waters • How to load it • Available Modules • Known Limitations • Where to Get More information 2 What is Python? • Modern programming language with a succinct and intuitive syntax • Has grown over the years to offer an extensive software ecosystem • Interprets JIT-compiled byte-code, but can take full advantage of AOT-compiled code • using python libraries with AOT code (numpy, scipy) • interfacing seamlessly with AOT code (CPython API) • generating native modules with/from AOT code (cython) 3 Why Python for HPC? • When you want to maximize productivity (not necessarily performance) • Mature language with large user base • Huge collection of freely available software libraries • High Performance Computing • Engineering, Optimization, Differential Equations • Scientific Datasets, Analysis, Visualization • General-purpose computing • web apps, GUIs, databases, and tons more • Python combines the best of both JIT and AOT code. • Write performance critical loops and kernels in C/FORTRAN • Write high level logic and “boiler plate” in Python 4 Python on Blue Waters • Blue Waters Python Software Stack (bw-python) • Over 60 python modules supporting HPC applications • Implements the Scipy Stack Specification (http://www.scipy.org/stackspec.html) • Provides extensions for parallel programming (mpi4py, pycuda) and scientific data IO (h5py, pycuda) • Numerical routines linked from ACML • Built specifically for running jobs on the compute nodes module swap PrgEnv-cray PrgEnv-gnu module load bw-python python ! Python 2.7.5 (default, Jun 2 2014, 02:41:21) [GCC 4.8.2 20131016 (Cray Inc.)] on Blue Waters Type "help", "copyright", "credits" or "license" for more information.