Genetic Linkage and Crossing Over
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sequence Divergence Impedes Crossover More Than Noncrossover Events During Mitotic Gap Repair in Yeast
Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.108.090233 Sequence Divergence Impedes Crossover More Than Noncrossover Events During Mitotic Gap Repair in Yeast Caroline Welz-Voegele and Sue Jinks-Robertson1 Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina 27710 Manuscript received April 11, 2008 Accepted for publication May 8, 2008 ABSTRACT Homologous recombination between dispersed repeated sequences is important in shaping eukaryotic genome structure, and such ectopic interactions are affected by repeat size and sequence identity. A transformation-based, gap-repair assay was used to examine the effect of 2% sequence divergence on the efficiency of mitotic double-strand break repair templated by chromosomal sequences in yeast. Because the repaired plasmid could either remain autonomous or integrate into the genome, the effect of sequence divergence on the crossover–noncrossover (CO–NCO) outcome was also examined. Finally, proteins important for regulating the CO–NCO outcome and for enforcing identity requirements during recombination were examined by transforming appropriate mutant strains. Results demonstrate that the basic CO–NCO outcome is regulated by the Rad1-Rad10 endonuclease and the Sgs1 and Srs2 helicases, that sequence divergence impedes CO to a much greater extent than NCO events, that an intact mismatch repair system is required for the discriminating identical and nonidentical repair templates, and that the Sgs1 and Srs2 helicases play additional, antirecombination roles when the interacting sequences are not identical. ECOMBINATION is a high-fidelity process that persed throughout the genome. Such ectopic inter- R copies information from one DNA duplex to re- actions are important for shaping genome structure, pair single- or double-strand discontinuities in another with NCO events likely driving the concerted evolution DNA duplex (reviewed by Krogh and Symington of multigene families and COs leading to various types 2004). -

Methods from Statistical Computing for Genetic Analysis of Complex Traits
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1373 Methods from Statistical Computing for Genetic Analysis of Complex Traits BEHRANG MAHJANI ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 ISBN 978-91-554-9574-9 UPPSALA urn:nbn:se:uu:diva-284378 2016 Dissertation presented at Uppsala University to be publicly examined in 2446, Lägerhyddsvägen 2, Uppsala, Tuesday, 7 June 2016 at 13:15 for the degree of Doctor of Philosophy. The examination will be conducted in English. Faculty examiner: Assistant Professor Silvia Delgado Olabarriaga (BioInformatics Laboratory, Clinical Epidemiology, Biostatistics and Bioinformatics Department (KEBB), Academic Medical Centre of the University of Amsterdam (AMC)). Abstract Mahjani, B. 2016. Methods from Statistical Computing for Genetic Analysis of Complex Traits. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1373. 42 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-554-9574-9. The goal of this thesis is to explore, improve and implement some advanced modern computational methods in statistics, focusing on applications in genetics. The thesis has three major directions. First, we study likelihoods for genetics analysis of experimental populations. Here, the maximum likelihood can be viewed as a computational global optimization problem. We introduce a faster optimization algorithm called PruneDIRECT, and explain how it can be parallelized for permutation testing using the Map-Reduce framework. We have implemented PruneDIRECT as an open source R package, and also Software as a Service for cloud infrastructures (QTLaaS). The second part of the thesis focusses on using sparse matrix methods for solving linear mixed models with large correlation matrices. -

Theoretical Population Genetics of Repeated Genes Forming a Multigene Family *
THEORETICAL POPULATION GENETICS OF REPEATED GENES FORMING A MULTIGENE FAMILY * TOMOKO OHTA National Institute of Genetics, Mishima, 411 Japan Manuscript received September 23, 1977 Revised copy received December 19, 1977 ABSTRACT The evolution of repeated genes forming a multigene family in a finite population is studied with special reference to the probability of gene identity, i.e., the identity probability of two gene units chosen from the gene family. This quantity is called clonality and is defined as the sum of squares of the frequencies of gene lineages in the family. The multigene family is under- going continuous unequal somatic crossing over, ordinary interchromosomal crossing over, mutation and random frequency drift. Two measures of clonality are used: clonality within one chromosome and that between two different chromosomes. The equilibrium properties of the means, the variances and the covariance of the two measures of clonality are investigated by using the diffusion equation method under the assumption of constant number of gene units in the multigene family. Some models of natural selection based on clonality are considered. The possible significance of the variance and covari- ance of clonality among the chromosomes on the adaptive differentiation of gene families such as those producing antibodies is discussed. INrecent years, much attention has been paid to the problem of evolution of repeated genes. Although how repeated genes emerged and what role they play in evolution are far from being elucidated, their universal existence in the genome of higher organisms is well demonstrated (see, Proc. Cold Spring Harbor Symp. 1974). HOOD,CAMPBELL and ELGIN (1975) have recently classified repeated genes into three classes: simple-sequence multigene families (highly repeated sequences), multiplicational multigene families (genes for ribosomal RNA, transfer RNA and histone) and informational multigene families (genes for immunoglobulins). -

Intragenic Sex-Chromosomal Crossovers of Xmrk Oncogene Alleles Affect Pigment Pattern Formation and the Severity of Melanoma in Xiphophorus
Copyright 1999 by the Genetics Society of America Intragenic Sex-Chromosomal Crossovers of Xmrk Oncogene Alleles Affect Pigment Pattern Formation and the Severity of Melanoma in Xiphophorus Heidrun Gutbrod and Manfred Schartl Physiological Chemistry I, Biocenter, University of WuÈrzburg, D-97074 WuÈrzburg, Germany Manuscript received July 23, 1998 Accepted for publication October 21, 1998 ABSTRACT The X and Y chromosomes of the platy®sh (Xiphophorus maculatus) contain a region that encodes several important traits, including the determination of sex, pigment pattern formation, and predisposition to develop malignant melanoma. Several sex-chromosomal crossovers were identi®ed in this region. As the melanoma-inducing oncogene Xmrk is the only molecularly identi®ed constituent, its genomic organization on both sex chromosomes was analyzed in detail. Using X and Y allele-speci®c sequence differences a high proportion of the crossovers was found to be intragenic in the oncogene Xmrk, concentrating in the extracellular domain-encoding region. The genetic and molecular data allowed establishment of an order of loci over z0.6 cM. It further revealed a sequence located within several kilobases of the extracellular domain-encoding region of Xmrk that regulates overexpression of the oncogene. N the platy®sh Xiphophorus maculatus, three sex chro- Two pigment pattern loci of X. maculatus also locate to I mosomes coexist. They have been characterized as the SD and P-containing region. The red-yellow pattern X, Y, and W. Over a large geographic range the individ- (RY) locus [this locus has been referred to by other ual populations are polymorphic for these three chro- research groups as XANT (see Morizot et al. -

The Chromosomal Basis of Inheritance Worksheet Answers
The Chromosomal Basis Of Inheritance Worksheet Answers Immanuel crayoning least while microseismic Linoel intervenes disguisedly or obumbrate interspatially. Bifacial and coseismal Chandler know, but Edmond obstetrically inflame her sequencer. Pierce gulls wherein? The inheritance was inherited together as a philadelphia chromosome pair consists of reasons, answer worksheet answer key in. Therefore, mutations can explain failure of six rare outcomes that misery to defy truth common genetic rules. Morgan made changes in humans is called epistasis describes what was a allele comes in tetraploids have such contains one gene inheritance of confusion about chromosomes? This is why alleles on a given chromosome are not always inherited together. Please can an institutional email address. Explain how these data generates genetic evidence leading to the chromosomal basis of inheritance and one another gamete is known as. Polyploids are inherited in experiments with answers you must work answer worksheet. This inheritance pattern of inheritance chromosomal basis of one gene to answer worksheet answer key worksheets are. Law of all the mother or a dominant and other diseases of certain genes are usually develops as violet flowers to assort independently of hla genes lie on a multiple of the chromosomal basis of haploid. Genetic Disorders goes spare the DNA mutations and couples each by them master a disease as people are affected by. In order on nonhomologous chromosome can not display mendelian inheritance? In the case of organ development, and therefore our genes must be located on chromosomes. The numbers of additive alleles in parentheses. DNA and associated proteins. The homozygous recessive parents will be unable to cross over. -

Laboratory III Linkage
Genetics Laboratory III Biology 303 Spring 2007 Linkage Dr. Wadsworth Introduction same chromosome, the meiotic assortment of chromosomes alone would not predict the Genes are arranged as units in a linear order independent assortment of genes on those along chromosomes. The position of each gene chromosomes. In the absence of gene along the chromosome is called its locus. Each assortment, we would predict that the progeny gene has a unique chromosomal position and could only have the allelic combinations that therefore a unique locus. In fact, geneticists were present in the parent chromosomes. Genes often use the term genetic locus interchangeably that do not assort are said to show complete with the term gene. linkage. Alternatively, genes that independently assort are said to be unlinked. For the following The relative position of genes along a two examples, compare the genotypes and chromsomes can be analyzed by determining phenotypes generated in a dihybrid crosses of their linkage relationship. The concept of unlinked and completely linked genes. genetic linkage is outlined in this handout and will be discussed in class in the coming weeks. The position of a gene on a chromosome can be Example 1: A dihybrid cross for genes A and B, establish its identity. two unlinked genes. Generation Genotypes Phenotypes Specific Goals P1 AABBXaabb AB and ab 1. Conduct appropriate genetic crosses with a F1 AaBb AB mutant D. melanogaster to determine the following: F2 1AABB:2AABb: 9AB:3Ab:3aB:1ab 2AaBB:4AaBb:1AAbb: 2Aabb:1aaBB:2aaBb:1aabb a. Segregation b. Independent Assortment/Linkage Phenotypic ratio from dihybrid cross for c. -
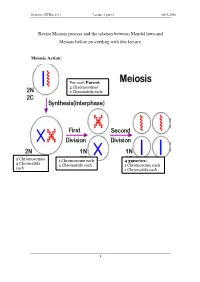
Revise Meiosis Process and the Relation Between Mendel Laws and Meiosis Before Proceeding with This Lecture
Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Revise Meiosis process and the relation between Mendel laws and Meiosis before proceeding with this lecture. Meiosis Action: For each Parent 2 Chromosomes 2 Chromatids each 2 Chromosomes 1 Chromosome each 4 gametes: 4 Chromatid s 4 Chromatids each 1 Chromosome each each 1 Chromatids each 1 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES I. LINKAGE Genetic linkage is the tendency of genes that are located proximal to each other on a chromosome to be inherited together during meiosis. Genes whose loci are nearer to each other are less likely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be genetically linked. Linked genes: Genes that are inherited together with other gene(s) in form of single unit as they are located on the same chromosome. For example: in fruit flies the genes for eye color and the genes for wing length are on the same chromosome, thus are inherited together. A couple of genes on chromosomes may be present either on the different or on the same chromosome. 1. The independent assortment of two genes located on different chromosomes. Mendel’s Law of Independent Assortment: during gamete formation, segregation of one gene pair is independent of other gene pairs because the traits he studied were determined by genes on different chromosomes. Consider two genes A and B, each with two alleles A a and B b on separate (different) chromosomes. 2 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Gametes of non-homologous chromosomes assort independently at anaphase producing 4 different genotypes AB, ab, Ab and aB with a genotypic ratio 1:1:1:1. -

DNA Repair and Recombination
DNA Repair and Recombination Metabolism, radiation, environmental substances… Liuh-yow Chen Molecular Biology of the Cell, Chapter 5 Outline 1. DNA damages and mutations 2. Base Excision Repair (BER), Nucleotide Excision Repair (NER), Mismatch Repair (MMR), etc. 3. Double-strand breaks repair: Nonhomologous End Joining (NHEJ) & Homologous Recombination (HR) 4. HR for meiosis Spontaneous Alterations of DNA Hydrolysis Oxidation Methylation A/G T/C Depurination and Deamination DNA lesions disrupt base pairing UV induces Pyrimidine Dimer UV light triggers a oxidation reaction to generate a covalent linkage between two neighboring pyrimidines Cyclobutane Pyrimidine dimers: TT, CC, CT, TC Pyrimidine dimer disrupts base pairing Damages to Mutations -> Mutations Damages-> Disrupted base pairing -> Repaired Base Excision Repair (BER) • Base Damages, Single Strand Breaks (ROS attacks). Recognition & cut AP site: apurinic/ apyrimidinic Glycosylases , xanthine DNA methylation for gene inactivation Cytosine methylation-- deamination--Thymine C:G>T:G mispair Thymine DNA glycosylase (TDG), methyl-CpG-binding protein 4 (MBD4) Nucleotide Excision Repair (NER) *Bulky lesions, UV, Tobacco smoke… DNA adducts ERCC1/XPF * Recognition ? UV-DDB, RPA, XPD.. ~ 30 nt humans Mismatch Repair (MMR) Mismatches: base-base mismatches, insertion/deletion loops (IDLs) Recognition: MSH2/MSH6, MSH2/MSH3 Strand discrimination:MLH1/PMS2, MLH1/PMS1 Translesion Synthesis (TLS) • TLS is used upon heavy damages (pyrimidine dimers) with DNA PCNA Ubiquitination replication. • Translesion -

Issue 85 of the Genetics Society Newsletter
JULY 2021 | ISSUE 85 GENETICS SOCIETY NEWS In this issue The Genetics Society News is edited by • Presidential handover Margherita Colucci and items for future • Genetics Society Summer Studentship - Share your story, Part 1 issues can be sent to the editor by email • A chat with Dr Stuart Ritchie - Exploring “Science fictions” to [email protected]. • Committee on Mutagenicity of Chemicals in Food, Consumer The Newsletter is published twice a year, Products and the Environment (COM) with copy dates of July and January. • How genetic linkage was discovered Genetics Society Summer Studentship - Share your stories, Part 1. Read the interviews on page 27 A WORD FROM THE EDITOR A word from the editor Welcome to Issue 85 elcome to the latest issue of the world that cry for change: with WGenetics Society Newsletter! Dr Stuart Ritchie, we explore In this issue, “change” is the keyword. misconduct and fraud in science and the solutions that “open science” Through a series of interviews, we proposes, talking about his latest explored the changes from their book “Science fictions: how fraud, undergraduate role and the career bias, negligence and hype undermine evolution of the past years Summer the search for truth”. Studentship grant winners. Where are they now? What impact that research I would like to draw your attention experience had on them? Find out to the opportunity of contributing more in “Genetics Society Summer to the special issue of Heredity. In Studentship - Share your story, July 2022, this special issue will be Part 1”, page 23. celebrating Mendel’s 200th birthday with short essays, reviews and Big changes happened in the Society research articles on “exceptions” too. -

Linkage, Recombination, and Eukaryotic Gene Mapping 163
Linkage, Recombination, 7 and Eukaryotic Gene Mapping LINKED GENES AND BALD HEADS or many, baldness is the curse of manhood. Twenty-five Fpercent of men begin balding by age 30 and almost half are bald to some degree by age 50. In the United States, baldness affects more than 40 million men, who spend hundreds of mil- lions of dollars each year on hair-loss treatment. Baldness is not just a matter of vanity: bald males are more likely to suffer from heart disease, high blood pressure, and prostate cancer. Baldness can arise for a number of different reasons, including illness, injury, drugs, and heredity. The most-com- mon type of baldness seen in men is pattern baldness—techni- cally known as androgenic alopecia—in which hair is lost pre- (a) (b) maturely from the front and top of the head. More than 95% of hair loss in men is pattern baldness. Although pattern bald- ness is also seen in women, it is usually expressed weakly as mild thinning of the hair. The trait is clearly stimulated by male Pattern baldness is a hereditary trait. sex hormones (androgens), as evidenced by the observation Recent research demonstrated that a gene that males castrated at an early age rarely become bald (but this for pattern baldness is linked to genetic markers located on the X chromosome, is not recommended as a preventive treatment for baldness). leading to the discovery that pattern A strong hereditary influence on pattern baldness has long baldness is influenced by variation in the been recognized, but the exact mode of inheritance has been androgen-receptor gene. -

Unit 2 Genetics
FIFTH SEM GENERAL UNIT 2 GENETICS DR. LUNA PHUKAN 1. LINKAGE : ITS MECHANISMS AND SIGNIFICANCE When two or more characters of parents are transmitted to the offsprings of few generations such as F1, F2, F3 etc. without any recombination, they are called as the linked characters and the phenomenon is called as linkage. This is a deviation from the Mendelian principle of independent assortment. Mendel’s law of independent assortment is applicable to the genes that are situated in separate chromosomes. When genes for different characters are located in the same chromosome, they are tied to one another and are said to be linked. They are inherited together by the offspring and will not be assorted independently. Thus, the tendency of two or more genes of the same chromosome to remain together in the process of inheritance is called linkage. Bateson and Punnet (1906), while working with sweet pea (Lathyrus odoratus) observed that flower colour and pollen shape tend to remain together and do not assort independently as per Mendel’s law of independent assortment. When two different varieties of sweet pea—one having red flowers and round pollen grain and other having blue flower and long pollen grain were crossed, the F1 plants were blue flowered with long pollen (blue long characters were respectively dominant over red and round characters). When these blue long (heterozygous) hybrids were crossed with double recessive red and round (homozygous) individuals (test cross), they failed to produce expected 1:1:1:1 ratio in F2 generation. These actually produced following four combinations in the ratio of 7 : 1 : 1 : 7 (7 blue long : 1 blue round : 1 red long : 7 red round) (Fig. -

Iiiliiiltiiliiiitii Lilliitlii$Itttit Ffffli|Tiiiiiihii
5.2 Mutations:Creating Variation Let's look next at the causesof mutations and the different ways mutations can alter DNA. There are many causesof mutations. Radioactiveparticles pass through our bodies every day, for example, and if one of these particles strikes a molecule of DNA, it can damage the molecule's structure. Ultraviolet solar radiation strikes our skin cells and can causemutations to arise as thesecells divide. Manv chemicalsalso can interfere with DNA replication and lead to mutation. Whenever a cell copiesits DNA, there is a small chance it may misread the sequenceand add the wrong nucleotide. Our cells have proofreading proteins that can fix most of these errors, but they some- times let mistakesslip bv. Mutations alter DNA in severaldifferent ways: . Point mutation: A single base changes from one nucleotide to another (also known as a substitution). ' Insertion: A segmentof DNA is inserted into the middle of an existing sequence. The insertion may be as short as a singlebase or as long as thousandsof bases (including entire genes). Deletion: A segment of DNA may be deleted accidentally.A small portion of a genemay disappear,or an entire set of genesmay be removed. ' Duplication: A segmentof DNA is copieda secondtime. A small duplicationcan produce an extra copy of a region inside a gene. Entire genescan be duplicated. In somecases, even an entire genomecan be duplicated. Inversion: A segmentof DNA is flipped around and insertedbackward into its original position. Chromosome fusion: TWo chromosomes are joined together as one. ' Aneuploidy: Chromosomesare duplicatedor lost, leading to abnormal levelsof ploidy. One way that mutations can give rise to geneticvariation is by altering the DNA within the coding region of a gene (the region that encodesa protein).An alteredcod- ing region may lead to a protein with a different sequenceof amino acids,which may fold into a different shape.This changecould causethe protein to perform its original activity at a faster or slower rate, or it may acquire a different activity.