Overexpression of LAMP3/TSC403/DC-LAMP Promotes Metastasis in Uterine Cervical Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ITGB6-Knockout Suppresses Cholangiocarcinoma Cell Migration and Invasion with Declining PODXL2 Expression
International Journal of Molecular Sciences Article ITGB6-Knockout Suppresses Cholangiocarcinoma Cell Migration and Invasion with Declining PODXL2 Expression Yurie Soejima 1,*,† , Miho Takeuchi 1, Nao Miyamoto 1, Motoji Sawabe 1 and Toshio Fukusato 2,† 1 Department of Molecular Pathology, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan; [email protected] (M.T.); [email protected] (N.M.); [email protected] (M.S.) 2 General Medical Education and Research Center, Teikyo University, 2-11-1 Kaga, Itabashi-ku, Tokyo 178-8605, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-3-5803-5375 † These authors contributed equally to this work. Abstract: Intrahepatic cholangiocarcinoma (iCCA) is a heterogeneous bile duct cancer with a poor prognosis. Integrin αvβ6 (β6) has been shown to be upregulated in iCCA and is associated with its subclassification and clinicopathological features. In the present study, two ITGB6-knockout HuCCT1 CCA cell lines (ITGB6-ko cells) were established using the clustered regulatory interspaced short palindromic repeats (CRISPR), an associated nuclease 9 (Cas9) system, and single-cell cloning. RNA sequencing analysis, real-time polymerase chain reaction (PCR), and immunofluorescent methods were applied to explore possible downstream factors. ITGB6-ko cells showed significantly decreased expression of integrin β6 on flow cytometric analysis. Both cell lines exhibited significant inhibition of cell migration and invasion, decreased wound-healing capability, decreased colony formation ability, and cell cycle dysregulation. RNA sequencing and real-time PCR analysis revealed a remarkable Citation: Soejima, Y.; Takeuchi, M.; decrease in podocalyxin-like protein 2 (PODXL2) expression in ITGB6-ko cells. -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Primary Myeloid Cell Proteomics and Transcriptomics
Primary myeloid cell proteomics and transcriptomics: importance of β-tubulin isotypes for osteoclast function David Guérit, Pauline Marie, Anne Morel, Justine Maurin, Christel Vérollet, Brigitte Raynaud-Messina, Serge Urbach, Anne Blangy To cite this version: David Guérit, Pauline Marie, Anne Morel, Justine Maurin, Christel Vérollet, et al.. Primary myeloid cell proteomics and transcriptomics: importance of β-tubulin isotypes for osteoclast function. Journal of Cell Science, Company of Biologists, 2020, 133 (10), pp.jcs239772. 10.1242/jcs.239772. hal- 02863726 HAL Id: hal-02863726 https://hal.archives-ouvertes.fr/hal-02863726 Submitted on 16 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Title Primary myeloid cell proteomics and transcriptomics: importance of ß tubulin isotypes for osteoclast function. Running title Primary myeloid cell omics and tubulin isotypes David Guérit1.2*. Pauline Marie1.2*. Anne Morel1.2. Justine Maurin1.2. Christel Verollet3.4. Brigitte Raynaud-Messina3.4. Serge Urbach5. Anne Blangy1.2@ 1 Centre de Recherche de Biologie Cellulaire de Montpellier (CRBM). CNRS UMR 5237. 1919 route de Mende. 34293 Montpellier cedex 5. France. 2 Montpellier University. 34095 Montpellier cedex 5. France. 3 Institut de Pharmacologie et Biologie Structurale. -

Novel Associations of BST1 and LAMP3 with Rapid Eye Movement Sleep Behavior
Published Ahead of Print on January 4, 2021 as 10.1212/WNL.0000000000011464 Neurology Publish Ahead of Print DOI: 10.1212/WNL.0000000000011464 Novel Associations of BST1 and LAMP3 with Rapid Eye Movement Sleep Behavior Disorder The Article Processing Charge was funded by NIHR UK This is an open access article distributed under the terms of the Creative Commons Attribution License 4.0 (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Neurology® Published Ahead of Print articles have been peer reviewed and accepted for publication. This manuscript will be published in its final form after copyediting, page composition, and review of proofs. Errors that could affect the content may be corrected during these processes. Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology. Kheireddin Mufti, MSc1,2, Eric Yu, MSc1,2, BSc, Uladzislau Rudakou, MSc1,2, Lynne Krohn, MSc1,2, Jennifer A. Ruskey, MSc2,3, Farnaz Asayesh, MSc2,3, Sandra B. Laurent, BTS2,3, Dan Spiegelman, MSc2,3, Isabelle Arnulf, MD, PhD4, Michele T.M. Hu, MBBS, PhD5,6, Jacques Y. Montplaisir, MD, PhD7,8, Jean-François Gagnon, PhD7,9, Alex Desautels, MD, PhD7,10, Yves Dauvilliers, MD, PhD11, Gian Luigi Gigli, MD12,13, Mariarosaria Valente, MD12,14, Francesco Janes, MD, PhD12, Andrea Bernardini, MD12, Birgit Högl, MD15, Ambra Stefani, MD15, Evi Holzknecht, MD15, Karel Sonka, MD, PhD16, David Kemlink, MD, PhD16, Wolfgang Oertel, MD17, Annette Janzen, MD17, Giuseppe Plazzi, MD18,19, Elena Antelmi, MD, PhD20, Michela Figorilli, MD, PhD21, Monica Puligheddu, MD, PhD21, Brit Mollenhauer, MD22,23, Claudia Trenkwalder, MD22.23, Friederike Sixel-Döring, MD17,22, Valérie Cochen De Cock, MD, PhD24,25 Christelle Charley Monaca, MD,PhD26, Anna Heidbreder, MD27, Luigi Ferini-Strambi, MD28, Femke Dijkstra, MD29,30,31, Mineke Viaene, MD, PhD29,30, Beatriz Abril, MD32, Bradley F. -

Single-Cell Analysis of Crohn's Disease Lesions Identifies
bioRxiv preprint doi: https://doi.org/10.1101/503102; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy JC Martin1,2,3, G Boschetti1,2,3, C Chang1,2,3, R Ungaro4, M Giri5, LS Chuang5, S Nayar5, A Greenstein6, M. Dubinsky7, L Walker1,2,5,8, A Leader1,2,3, JS Fine9, CE Whitehurst9, L Mbow9, S Kugathasan10, L.A. Denson11, J.Hyams12, JR Friedman13, P Desai13, HM Ko14, I Laface1,2,8, Guray Akturk1,2,8, EE Schadt15,16, S Gnjatic1,2,8, A Rahman1,2,5,8, , M Merad1,2,3,8,17,18*, JH Cho5,17,*, E Kenigsberg1,15,16,17* 1 Precision Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 2 Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 3 Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 4 The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York City, NY 10029, USA. 5 Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 6 Department of Colorectal Surgery, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA 7 Department of Pediatrics, Susan and Leonard Feinstein IBD Clinical Center, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -

Sensitization to the Lysosomal Cell Death Pathway by Oncogene- Induced Down-Regulation of Lysosome-Associated Membrane Proteins 1 and 2
Research Article Sensitization to the Lysosomal Cell Death Pathway by Oncogene- Induced Down-regulation of Lysosome-Associated Membrane Proteins 1 and 2 Nicole Fehrenbacher,1 Lone Bastholm,2 Thomas Kirkegaard-Sørensen,1 Bo Rafn,1 Trine Bøttzauw,1 Christina Nielsen,1 Ekkehard Weber,3 Senji Shirasawa,4 Tuula Kallunki,1 and Marja Ja¨a¨ttela¨1 1Apoptosis Department and Centre for Genotoxic Stress Response, Institute for Cancer Biology, Danish Cancer Society; 2Institute of Molecular Pathology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark; 3Institute of Physiological Chemistry, Medical Faculty, Martin-Luther-University Halle-Wittenberg, Halle, Germany; and 4Department of Cell Biology, School of Medicine, Fukuoka University, Fukuoka, Japan Abstract molecules of the cell to breakdown products available for Expression and activity of lysosomal cysteine cathepsins metabolic reuse (3, 4). Cathepsin proteases are among the best- correlate with the metastatic capacity and aggressiveness of studied lysosomal hydrolases. They are maximally active at the tumors. Here, we show that transformation of murine acidic pH of lysosomes (pH 4–5). However, many of them can be Y527F embryonic fibroblasts with v-H-ras or c-src changes the active at the neutral pH outside lysosomes, albeit with a decreased distribution, density, and ultrastructure of the lysosomes, efficacy and/or altered specificity (5). For example, transformation and tumor environment enhance the expression of lysosomal decreases the levels of lysosome-associated membrane pro- teins (LAMP-1 and LAMP-2) in an extracellular signal- cysteine cathepsins and increase their secretion into the extracel- regulated kinase (ERK)- and cathepsin-dependent manner, lular space (6). Once outside the tumor cells, cathepsins stimulate and sensitizes the cells to lysosomal cell death pathways angiogenesis, tumor growth, and invasion in murine cancer induced by various anticancer drugs (i.e., cisplatin, etoposide, models, thereby enhancing cancer progression (7, 8). -

Original Article Expression of LAMP3 and Its Correlation with Clinicopathologic Characteristics and Prognosis in Hepatocellular Carcinoma
Int J Clin Exp Pathol 2018;11(1):367-374 www.ijcep.com /ISSN:1936-2625/IJCEP0067961 Original Article Expression of LAMP3 and its correlation with clinicopathologic characteristics and prognosis in hepatocellular carcinoma Yang Gui1,2*, Wen-Bin Liu1,2*, Hao Chen1,2, Jin-Liang Ma1,2, Jian-Sheng Li1,2 1Department of General Surgery, Affiliated Provincial Hospital of Anhui Medical University, Hefei, Anhui, China; 2Anhui Province Key Laboratory of Hepatopancreatobiliary Surgery, Hefei, Anhui, China. *Equal contributors. Received October 26, 2017; Accepted November 10, 2017; Epub January 1, 2018; Published January 15, 2018 Abstract: Lysosome-associated membrane protein (LAMP) 3 is one of members of lysosome-associated membrane protein family, which has been reported to play an important role in multiple malignant tumors. However, there is less research about the expression of LAMP3 in hepatocellular carcinoma (HCC). The purpose of this study was to investigate the expression of LAMP3 and explore its roles in HCC.The expression of LAMP3 in 99 cases of HCC tissues was performed by immunohistochemistry. In addition, the expression of LAMP3 in 20 pairs of HCC tissues and pericarcinomatous tissues was determined by quantitative real-time polymerase chain reaction and Western blotting. Immunohistochemical staining showed that LAMP3 was mainly expressed in the cytoplasm. And the ex- pression of LAMP3 in HCC tissues (64/99, 64.6%) was significantly lower than that in pericarcinomatous tissues (23/99, 23.2%). In addition, the expression of LAMP3 mRNA and protein in HCC tissues was also significantly lower than that in pericarcinomatous tissues for 20 pairs of HCC samples. Low expression of LAMP3 was correlated with age, tumor-node-metastasis (TNM) staging, Edmondson grade, alpha-fetoprotein (AFP). -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
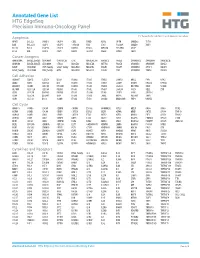
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Mouse CD Marker Chart Bdbiosciences.Com/Cdmarkers
BD Mouse CD Marker Chart bdbiosciences.com/cdmarkers 23-12400-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1d CD1.1, CD1.2, Ly-38 Lipid, Glycolipid Ag + + + + + + + + CD104 Integrin b4 Laminin, Plectin + DNAX accessory molecule 1 (DNAM-1), Platelet and T cell CD226 activation antigen 1 (PTA-1), T lineage-specific activation antigen 1 CD112, CD155, LFA-1 + + + + + – + – – CD2 LFA-2, Ly-37, Ly37 CD48, CD58, CD59, CD15 + + + + + CD105 Endoglin TGF-b + + antigen (TLiSA1) Mucin 1 (MUC1, MUC-1), DF3 antigen, H23 antigen, PUM, PEM, CD227 CD54, CD169, Selectins; Grb2, β-Catenin, GSK-3β CD3g CD3g, CD3 g chain, T3g TCR complex + CD106 VCAM-1 VLA-4 + + EMA, Tumor-associated mucin, Episialin + + + + + + Melanotransferrin (MT, MTF1), p97 Melanoma antigen CD3d CD3d, CD3 d chain, T3d TCR complex + CD107a LAMP-1 Collagen, Laminin, Fibronectin + + + CD228 Iron, Plasminogen, pro-UPA (p97, MAP97), Mfi2, gp95 + + CD3e CD3e, CD3 e chain, CD3, T3e TCR complex + + CD107b LAMP-2, LGP-96, LAMP-B + + Lymphocyte antigen 9 (Ly9), -

Applies to Article Number 95311, Released June 13, 2019
Applies to article number 95311, released June 13, 2019 Adenosine deaminase (ADA) P00813 Decorin (DCN) P07585 Adhesion G-protein coupled receptor G1 (ADGRG1) Q9Y653 Fibroblast growth factor 2 (FGF2) P09038 Angiopoietin-1 (ANGPT1) Q15389 Fractalkine (CX3CL1) P78423 Angiopoietin-1 receptor (TIE2) Q02763 Galectin-1 (Gal-1) P09382 Angiopoietin-2 (ANGPT2) O15123 Galectin-9 (Gal-9) O00182 Arginase-1 (ARG1) P05089 Granzyme A (GZMA) P12544 Carbonic anhydrase 9 (CAIX) Q16790 Granzyme B (GZMB) P10144 Caspase-8 (CASP-8) Q14790 Granzyme H (GZMH) P20718 C-C motif chemokine 13 (MCP-4) Q99616 Heme oxygenase 1 (HO-1) P09601 C-C motif chemokine 17 (CCL17) Q92583 Hepatocyte growth factor (HGF) P14210 C-C motif chemokine 19 (CCL19) Q99731 ICOS ligand (ICOSLG) O75144 C-C motif chemokine 2 (MCP-1) P13500 Interferon gamma (IFN-gamma) P01579 C-C motif chemokine 20 (CCL20) P78556 Interleukin-1 alpha (IL-1 alpha) P01583 C-C motif chemokine 23 (CCL23) P55773 Interleukin-10 (IL10) P22301 C-C motif chemokine 3 (CCL3) P10147 Interleukin-12 (IL12) P29459, P29460 C-C motif chemokine 4 (CCL4) P13236 Interleukin-12 receptor subunit beta-1 (IL12RB1) P42701 C-C motif chemokine 7 (MCP-3) P80098 Interleukin-13 (IL13) P35225 C-C motif chemokine 8 (MCP-2) P80075 Interleukin-15 (IL15) P40933 CD27 antigen (CD27) P26842 Interleukin-18 (IL18) Q14116 CD40 ligand (CD40-L) P29965 Interleukin-2 (IL2) P60568 CD40L receptor (CD40) P25942 Interleukin-33 (IL33) O95760 CD70 antigen (CD70) P32970 Interleukin-4 (IL4) P05112 CD83 antigen (CD83) Q01151 Interleukin-5 (IL5) P05113 C-X-C motif -

Hypoxia Stimulates Migration of Breast Cancer Cells Via the PERK/ATF4
Nagelkerke et al. Breast Cancer Research 2013, 15:R2 http://breast-cancer-research.com/content/15/1/R2 RESEARCHARTICLE Open Access Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response Anika Nagelkerke1,2*, Johan Bussink1, Hilda Mujcic3,4, Bradly G Wouters3,4, Steffi Lehmann5, Fred CGJ Sweep2 and Paul N Span1,2 Abstract Introduction: The hypoxia-inducible factor (HIF)-1 pathway can stimulate tumor cell migration and metastasis. Furthermore, hypoxic tumors are associated with a poor prognosis. Besides the HIF-1 pathway, the unfolded protein response (UPR) is also induced by hypoxic conditions. The PKR-like ER kinase (PERK)/activating transcription factor 4 (ATF4)-arm of the UPR induces expression of lysosomal-associated membrane protein 3 (LAMP3), a factor that has been linked to metastasis and poor prognosis in solid tumors. In this study the role of UPR-induced LAMP3 in hypoxia-mediated migration of breast cancer cells was examined. Methods: A number of in vitro metastasis models were used to study the migration and invasion of MDA-MB-231 breast cancer cells under hypoxic conditions. PERK, ATF4 and their downstream factor LAMP3 were knocked down to examine their role in cell migration. In addition, multicellular tumor spheroids were used to study the involvement of the tumor microenvironment in invasion. Results: Using transwell assays, migration of different breast cancer cell lines was assessed. A direct correlation was found between cell migration and baseline LAMP3 expression. Furthermore, moderate hypoxia (1% O2) was found to be optimal in stimulating migration of MDA-MB-231 cells.