Genomics Data 14 (2017) 91–105
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Case Study of the Exotic Peregrine Earthworm Morphospecies Pontoscolex Corethrurus Shabnam Taheri, Céline Pelosi, Lise Dupont
Harmful or useful? A case study of the exotic peregrine earthworm morphospecies Pontoscolex corethrurus Shabnam Taheri, Céline Pelosi, Lise Dupont To cite this version: Shabnam Taheri, Céline Pelosi, Lise Dupont. Harmful or useful? A case study of the exotic peregrine earthworm morphospecies Pontoscolex corethrurus. Soil Biology and Biochemistry, Elsevier, 2018, 116, pp.277-289. 10.1016/j.soilbio.2017.10.030. hal-01628085 HAL Id: hal-01628085 https://hal.archives-ouvertes.fr/hal-01628085 Submitted on 5 Jan 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Harmful or useful? A case study of the exotic peregrine earthworm MARK morphospecies Pontoscolex corethrurus ∗ ∗∗ S. Taheria, , C. Pelosib, L. Duponta, a Université Paris Est Créteil, Université Pierre et Marie Curie, CNRS, INRA, IRD, Université Paris-Diderot, Institut d’écologie et des Sciences de l'environnement de Paris (iEES-Paris), Créteil, France b UMR ECOSYS, INRA, AgroParisTech, Université Paris-Saclay, 78026 Versailles, France ABSTRACT Exotic peregrine earthworms are often considered to cause environmental harm and to have a negative impact on native species, but, as ecosystem engineers, they enhance soil physical properties. Pontoscolex corethrurus is by far the most studied morphospecies and is also the most widespread in tropical areas. -

Natural Resources Research Institute, University of Minnesota Duluth
This document is made available electronically by the Minnesota Legislative Reference Library as part of an ongoing digital archiving project. http://www.leg.state.mn.us/lrl/lrl.asp 2009 Project Abstract For the Period Ending December 30, 2012 PROJECT TITLE: Prevention and Early Detection of Asian Earthworms and Reducing the Spread of European Earthworms PROJECT MANAGER: Cindy Hale AFFILIATION: Natural Resources Research Institute, University of Minnesota Duluth MAILING ADDRESS: 5013 Miller Trunk Hwy CITY/STATE/ZIP: Duluth MN 55811 PHONE: 218/720-4364 E-MAIL: [email protected] WEBSITE: [If applicable] FUNDING SOURCE: Environment and Natural Resources Trust Fund LEGAL CITATION: http://www.nrri.umn.edu/staff/chale.asp APPROPRIATION AMOUNT: $150,000 Overall Project Outcome and Results We used a multi-pronged approach to quantify of the relative importance of different vectors of spread for invasive earthworms, make management and regulatory recommendations and create mechanisms for public engagement and dissemination of our project results through the Great Lakes Worm Watch website and diverse stakeholders. Internet sales of earthworms and earthworm related products posed large risks for the introduction of new earthworm species and continued spread of those already in the state. Of 38 earthworm products sampled, 87% were either contaminated with other earthworm species or provided inaccurate identification. Assessment of soil transported via ATV’s and logging equipment demonstrated that this is also a high risk vector for spread of earthworms across the landscape, suggesting that equipment hygiene, land management activities and policies should address this risk. Preliminary recommendations for organizations with regulatory oversight for invasive earthworms (i.e. -

F:\Zoos'p~1\2004\March2~1
CASE REPORT ZOOS' PRINT JOURNAL 19(3): 1394-1400 ILLUSTRATIONS OF EARTHWORMS OCCURING IN AND AROUND CHENNAI, INDIA V.I. Ramzan Begum 1 and Sultan Ahmed Ismail 2 1 Institute of Research in Soil Biology and Biotechnology, The New College, Chennai, Tamil Nadu 600014, India. 2 Corresponding author: Ecoscience Research Foundation, Plot 98, Baaz Nagar, 3/621 East Coast Road, Palavakkam, Chennai, Tamil Nadu 600041, India. 2 Email: [email protected] Abstract made to keep the description simple. Earthworms were collected from Chennai and its environs for taxonomic listing and to prepare a brief guide for Earthworms occur all over the world, but only rarely in deserts, identification by budding vermiculturists. Earthworm areas under constant snow and ice, mountain ranges and areas species were identified using existing taxonomic keys. almost entirely lacking in soil and vegetation (Edwards & Bohlen, Drawida scandens, Lampito mauritii, Perionyx 1996). excavatus, Perionyx sansibaricus, Octochaetona barnesi, Octochaetona pattoni, Octochaetona serrata, Earthworms belong to the Order Oligochaeta of the Phylum Octochaetona thurstoni, Dichogaster bolaui and Annelida. Oligochaetes are bilaterally symmetrical coelomate Eudrilus eugeniae have been recorded. D. bolaui was invertebrates with internal and external metameric segmentation the smallest while O. thurstoni was the largest species in throughout the body. Oligochaetes are often divided into two size observed. Fully mature adults of the common species convenient groups Microdrili and Megadrili. During the last collected over a period of time from sites in and around decade and half (Ismail, 1997) much taxonomic work on Indian Chennai are illustrated. Among the species, Eudrilus earthworms has been carried out by Julka (1975, 1976a,b, 1977, eugeniae does not occur naturally and are procured from 1978, 1979, 1981, 1983, 1993), Jamieson (1977) and Easton (1982). -
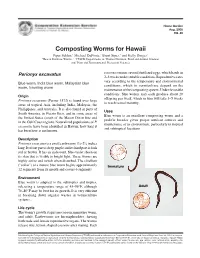
Composting Worms for Hawaii
Home Garden Aug. 2005 HG-46 Composting Worms for Hawaii Piper Selden,1 Michael DuPonte,2 Brent Sipes,3 and Kelly Dinges2 1Hawaii Rainbow Worms, 2, 3CTAHR Departments of 2Human Nutrition, Food and Animal Sciences and 3Plant and Environmental Protection Sciences Perionyx excavatus cocoon contains several fertilized eggs, which hatch in 2–3 weeks under suitable conditions. Reproductive rates Blue worm, India blue worm, Malaysian blue vary according to the temperature and environmental worm, traveling worm conditions, which in vermiculture depend on the maintenance of the composting system. Under favorable Origin conditions, blue worms may each produce about 20 offspring per week, which in turn will take 3–5 weeks Perionyx excavatus (Perrier 1872) is found over large to reach sexual maturity. areas of tropical Asia, including India, Malaysia, the Phillippines, and Australia. It is also found in parts of Uses South America, in Puerto Rico, and in some areas of Blue worm is an excellent composting worm and a the United States (south of the Mason Dixon line and prolific breeder given proper nutrient sources and in the Gulf Coast region). Naturalized populations of P. maintenance of its environment, particularly in tropical excavatus have been identified in Hawaii; how long it and subtropical locations. has been here is not known. Description 1 3 Perionyx excavatus is a small earthworm 1 ⁄4–2 ⁄4 inches long. Its front part is deep purple and its hind part is dark red or brown. It has an iridescent, blue-violet sheen on its skin that is visible in bright light. These worms are highly active and twitch when disturbed. -

Phylogenetic and Phenetic Systematics of The
195 PHYLOGENETICAND PHENETICSYSTEMATICS OF THE OPISTHOP0ROUSOLIGOCHAETA (ANNELIDA: CLITELLATA) B.G.M. Janieson Departnent of Zoology University of Queensland Brisbane, Australia 4067 Received September20, L977 ABSTMCT: The nethods of Hennig for deducing phylogeny have been adapted for computer and a phylogran has been constructed together with a stereo- phylogran utilizing principle coordinates, for alL farnilies of opisthopor- ous oligochaetes, that is, the Oligochaeta with the exception of the Lunbriculida and Tubificina. A phenogran based on the sane attributes conpares unfavourably with the phyLogralnsin establishing an acceptable classification., Hennigrs principle that sister-groups be given equal rank has not been followed for every group to avoid elevation of the more plesionorph, basal cLades to inacceptabl.y high ranks, the 0ligochaeta being retained as a Subclass of the class Clitellata. Three orders are recognized: the LumbricuLida and Tubificida, which were not conputed and the affinities of which require further investigation, and the Haplotaxida, computed. The Order Haplotaxida corresponds preciseLy with the Suborder Opisthopora of Michaelsen or the Sectio Diplotesticulata of Yanaguchi. Four suborders of the Haplotaxida are recognized, the Haplotaxina, Alluroidina, Monil.igastrina and Lunbricina. The Haplotaxina and Monili- gastrina retain each a single superfanily and fanily. The Alluroidina contains the superfamiJ.y All"uroidoidea with the fanilies Alluroididae and Syngenodrilidae. The Lurnbricina consists of five superfaniLies. -

Program Book
Table of Contents 66th State Science and Engineering Fair of Florida FFFS Purpose and Objectives ……………………………………...….…….........…...2 FFFS Board of Directors ………………………………………..……......……..…......3 SSEF History ……………………………………....................................................4 – 5 RSEF Directors …………….………………….....………………………......…...…...6 Scholarships and Opportunity Awards …………………...…………..............……7 – 8 Special and Premium Awards………………………...…………..................……9 – 14 Finalists and Projects ………….………………...…………………....…....…....15 – 42 Finalists Index ………………………………………….....………….....……….43 – 47 Category Judges ………………………………….....………..…………......…..48 – 50 A FOUND ID AT R IO O L N F F O S Ye6ars of Excellence T R S I F SSEF T U N T IE URE SC 1 Florida Foundation for Future Scientists FFFS Purpose and Objectives FFFSFFFS Purpose Purpose and and Objectives Objectives The Florida Foundation for Future Scientists (FFFS) is a statewide, non-profit organization authorized by the The Florida Foundation for Future Scientists (FFFS) is a statewide, non-profit organization authorized by the 1957 Legislature of the State of Florida to discover scientific and technical talent in the schools of Florida and to 1957 Legislature of the State of Florida to discover scientific and technical talent in the schools of Florida and to encourage the pursuit of careers in STEM (science, technology, engineering, and math). In meeting its encourage the pursuit of careers in STEM (science, technology, engineering, and math). In meeting its obligations, the FFFS promotes -

The Role of Black Soldier Fly, Hermetia Illucens (L.) (Diptera: Stratiomyidae) in Sustainable Waste Management in Northern Climates
University of Windsor Scholarship at UWindsor Electronic Theses and Dissertations Theses, Dissertations, and Major Papers 2012 The Role of Black Soldier Fly, Hermetia illucens (L.) (Diptera: Stratiomyidae) in Sustainable Waste Management in Northern Climates Luis Alvarez University of Windsor Follow this and additional works at: https://scholar.uwindsor.ca/etd Recommended Citation Alvarez, Luis, "The Role of Black Soldier Fly, Hermetia illucens (L.) (Diptera: Stratiomyidae) in Sustainable Waste Management in Northern Climates" (2012). Electronic Theses and Dissertations. 402. https://scholar.uwindsor.ca/etd/402 This online database contains the full-text of PhD dissertations and Masters’ theses of University of Windsor students from 1954 forward. These documents are made available for personal study and research purposes only, in accordance with the Canadian Copyright Act and the Creative Commons license—CC BY-NC-ND (Attribution, Non-Commercial, No Derivative Works). Under this license, works must always be attributed to the copyright holder (original author), cannot be used for any commercial purposes, and may not be altered. Any other use would require the permission of the copyright holder. Students may inquire about withdrawing their dissertation and/or thesis from this database. For additional inquiries, please contact the repository administrator via email ([email protected]) or by telephone at 519-253-3000ext. 3208. The Role of Black Soldier Fly, Hermetia illucens (L.) (Diptera: Stratiomyidae) in Sustainable Waste Management in Northern Climates by Luis Alvarez M.A.Sc., P.Eng. A Dissertation Submitted to the Faculty of Graduate Studies through Civil and Environmental Engineering in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy at the University of Windsor Windsor, Ontario, Canada 2012 © 2012 Luis Alvarez The Role of Black Soldier Fly, Hermetia illucens (L.) (Diptera: Stratiomyidae) in Sustainable Waste Management in Northern Climates by Luis Alvarez APPROVED BY: __________________________________________________ Dr. -

Cultivation of Eisenia Andrei Earthworms for Use As Feed for Aquatic Organisms
Available online at www.worldscientificnews.com WSN 152 (2021) 39-54 EISSN 2392-2192 Organic permaculture: cultivation of Eisenia andrei earthworms for use as feed for aquatic organisms Selivanov Yevhen, Marenkov Oleh* Department of General Biology and Water Bioresources, Faculty of Biology and Ecology, Oles Honchar Dnipro National University, P.M.B. 49050, Dnipro, Ukraine *E-mail address: [email protected] , [email protected] ABSTRACT This paper examines the biological features and taxonomic status of the red wiggler worm Eisenia andrei as presented in the relevant literature. We evaluate the economic feasibility of cultivating this species as feed for aquaculture purposes and discuss conventional cultivation methods. Keywords: Eisenia andrei, Eisenia fetida, aquaculture, vermiculture, feed 1. INTRODUCTION The modern approach in organic agriculture development relies on the scientific design of the production environment, as well as farming optimization based on natural interconnections between ecosystems. This approach is called "permaculture" (a portmanteau of "permanent agriculture"). The aim of permaculture development is focused on the creation of self-sustaining closed biosystems for agricultural production (fishery, aquaculture, animal husbandry, etc.) while utilizing traditional agricultural methods along with modern advances in science and technology. The development of permaculture includes the development of aquaculture as one of its key areas. Aquaculture is a controlled process of farming aquatic organisms, including fish, mollusks, crustaceans, and aquatic plants [1]. In terms of production, aquaculture has ( Received 21 November 2020; Accepted 05 December 2020; Date of Publication 06 December 2020 ) World Scientific News 152 (2021) 39-54 surpassed capture fisheries and is growing faster than any other branch of the food industry; in 2018, about 46% of the global aquatic animal production was aquaculture-sourced [1, 2]. -

Biomolecular Approach to Oligochaete Taxonomy Dr
Dr. Jaya M et. al. / International Journal of New Technologies in Science and Engineering Vol. 2, Issue 6,Dec 2015, ISSN 2349-0780 Biomolecular Approach To Oligochaete Taxonomy Dr. Jaya. M 1* ,Dr. Aja. M2 and Dr. K. Vijayakumaran Nair3 1* Assistant Professor, Department of Zoology, Sree keralavarma College, Thrissur, Email: [email protected] 2Senior Research Fellow, Department of Zoology, University of Kerala, Kariavatom, Email: [email protected] 3Associate Professor, Department of Zoology, Mar Ivanios College, Thiruvananthapuram, Email: [email protected] ABSTRACT This paper comprises the molecular approach for the identification of earthworm along with the traditional taxonomic method. The mitochondrial CO 1 gene of the Pontoscolex corethrurus (Glossoscolecidae), Travoscolides chengannures, Amynthas corticis, Perionyx sansibaricus (Megascolecidae), Progizzardus varadiamensis and Glyphidrilus annandalei (Almidae) were sequenced. The cytochrome-c oxidase I (CO1) exhibited a unique barcode to a particular species. The further exploration of mitochondrial diversity in earthworms will lead to major improvements in our understanding of the evolutionary pathways and rates of the mitochondrial genome Key words: Barcoding, cytochrome c oxidase 1 (COI), 16S ribosomal DNA INTRODUCTION DNA barcoding is a taxonomic method that uses a short genetic marker in the DNA to identify an organism. It differs from molecular phylogeny in that the main goal is to identify an unknown sample in terms of a known classification [26]. DNA sequence can be used to identify different species, in the same way as the supermarket scanner uses the black stripes of the UPC barcode to identify the items. This database will rapidly link a specimen to a Binomial Linnaean name and through that link it will provide all available information and studies on the species. -

What's Wrong with Worms?
WHAT’S WRONG WITH WORMS? Clay Antieau MS PhC Botanist, Horticulturist Environmental Educator Seventh Western Native Plants Conference, December 2016 EARTHWORM DIGESTIVE SYSTEM (Horn, Schramm, and Drake 2003) • Live 4 to 15+ years • Eats their weight in soil/organic matter daily • Food is processed in intestine (alimentary canal) Muscular mixing with enzymes and microbes in gut (anoxic) to release amino acids, sugars, organic molecules, nitrogen…. Molecules absorbed through intestinal membranes • Waste Product: CASTINGS BENEFITS OF EARTHWORMS (OM/soil digestion and bioturbation) • Improve soil physical structure better drainage/infiltration and aeration reduced stormwater run-off improved root penetration • Decompose, Mineralize OM Concentrate carbon and nutrients Enhanced soil fertility and tilth for plant growth, seed germination, crop yield. NUTRIENT VALUE OF CASTINGS • Nitrogen 1.80–2.05% • Phosphorus 1.32–1.93% • Potassium 1.28–1.50% • Calcium 3.0–4.5% • Magnesium 0.4–0.7% • Iron 0.3–0.7% • Manganese traces–0.40% • Zinc 0.028–0.036% • Organic Carbon 20-30% • pH 6.0–7.0 Darwin Earthworms! “The plow is one of the most ancient and most valuable of Man’s inventions; but long before he existed, the land … was regularly ploughed, and still continues to be ploughed, by earthworms. It may be (doubtful) whether there are many other animals which have played so important a part in the history of the world as these lowly, organized creatures.” - Charles Darwin (Earthworms 1881) …his last book, published just six months before he died. Different earthworms… Different functions…. Compost Dwellers (Epigeic) Live in high organic matter environments Eisenia fetida (red wriggler; tiger worm) Soil Surface Dwellers (Epigeic) Feed on decaying roots, shoots, leaves, and dung and live on or near soil surface (0-15 cm depth) Lumbricus rubellus (European earthworm; red wriggler) Topsoil Dwellers (Endogeic) Live in the top 20-30 cm of soil. -

Earthworm Ecology from DARWIN to VERMICUL TURE for 1882
Earthworm Ecology FROM DARWIN TO VERMICUL TURE FOR 1882. MAN ·I~ ·BVT ·A·woR...JV\· Frontispiece Cartoon from Punch, December 6th, 1881 Earthworm Ecology FROM DARWIN TO VERMICUL TURE Edited by J. E. Satchell Institute of Terrestrial Ecology Merlewood Research Station Grange-over-Sands Cumbria, UK LONDON NEW YORK CHAPMAN AND HALL First published 1983 by Chapman and Hall Ltd I I New Fetter Lane, London EC4P 4EE Published in the USA by Chapman and Hall 733 Third Avenue, New York NY100I7 © 1983 Chapman and Hall Ltd Softcover reprint of the hardcover 1st edition 2007 University Press, Cambridge ISBN-13: 978-94-009-5967-5 e-ISBN-13: 978-94-009-5965-1 DOl: 10.1007/978-94-009-5965-1 All rights reserved. No part ofthis book may be reprinted, or reproduced or utilized in any form or by any electronic, mechanical or other means, now known or hereafter invented, including photocopying and record ing, or in any information storage and retrieval system, without permission in writing from the Publisher. British Library Cataloguing in Publication Data Earthworm ecology. I. Opisthopora I. Satchell, J. E. 595.I' 46 QL39 I. 04 Library of Congress Cataloging in Publication Data Main entry under title: Earthworm ecology. Bibliography: p. Includes index. I. Opisthopora-Ecology. 2. Earthworm culture. I. Satchell, John E. QL39I.A6E2 7 1983 Contents Preface Xl Contributors xiii DARWIN'S CONTRIBUTION TO EARTHWORM ECOLOGY I Darwin's Formation of Vegetable Mould- its philo sophical basis M. S. Ghilarov I 2 D~rwin on earthworms - the contemporary back ground and what the critics thought O. -

Title of the Paper
Behera ‒ Patnaik: Negative effect of phosphogypsum over physiological activity of earthworm Eisenia fetida - 4455 - NEGATIVE EFFECT OF PHOSPHOGYPSUM OVER PHYSIOLOGICAL ACTIVITY OF EARTHWORM EISENIA FETIDA BEHERA, A. K.* – PATNAIK, A. School of Life Sciences, Sambalpur University, Jyoti Vihar, Burla, Odisha 768019, India (phone: +91-89-8423-7334) *Corresponding author e-mail: [email protected] (Received 22nd Mar 2018; accepted 27th Jun 2018) Abstract. The study aimed to assess the genuine impact of phosphogypsum on the growth, feeding, respiration and regeneration of earthworm Eisenia fetida. In laboratory condition the earthworms were cultured under 0% (control), 4%, 8% and 10% concentration of phosphogypsum for 30 days. After completion of every 10 day changes in the above parameters were observed to track the impact of phosphogypsum. With increasing exposure duration and concentration of phosphogypsum lower growth rate, declined feeding habit, maximum respiration rate and deprived regeneration power were noticed. The highest and lowest growths were 1.39 gm at 0% and 0.05 gm at 10%, respectively. Maximum feeding rate was 32.65 with a minimum rate 16.20 g g-1 live tissue. Respiration rate was highest at 10% i.e. 0.0578 g-1 live worm tissue hr-1 kg-1 soil, as most of the energy used to respire to sustain in such diverse condition and 0.575 g-1 live worm tissue hr-1 kg-1 soil recorded as lowest in 0%. The rate of regeneration was deeply hampered and there was no viable worms left at 8% and 10% concentration to assess. Regeneration was only observed at 0% and 4%.