Annual Report 2015 Rajiv Gandhi Centre for Biotechnology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Sl.No. APPL NO. Register No. APPLICANT NAME WITH
tpLtp vz;/ 7166 -2018-v Kjd;ik khtl;l ePjpkd;wk;. ntYhh;. ehs; 01/08/2018 mwptpf;if mytyf cjtpahsh; (Office Assistant) gzpfSf;fhd fPH;f;fhqk; kDjhuh;fspd; tpz;zg;g';fs; mLj;jfl;l eltof;iff;fhf Vw;Wf;bfhs;sg;gl;lJ/ nkYk; tUfpd;w 18/08/2018 kw;Wk; 19/08/2018 Mfpa njjpfspy; fPH;f;fz;l ml;ltizapy; Fwpg;gpl;Ls;s kDjhuh;fSf;F vGj;Jj; njh;t[ elj;j jpl;lkplg;gl;Ls;sJ/ njh;tpy; fye;Jbfhs;Sk; tpz;zg;gjhuh;fs; fPH;fz;l tHpKiwfis jtwhky; gpd;gw;wt[k;/ tHpKiwfs; 1/ njh;t[ vGj tUk; kDjhuh;fs; j’;fspd; milahs ml;il VnjDk; xd;W (Mjhu; ml;il - Xl;Leu; cupkk; - thf;fhsu; milahs ml;il-ntiytha;g;g[ mYtyf milahs ml;il) jtwhky; bfhz;Ltut[k;/ 2/ njh;t[ vGj tUk; kDjhuh;fs; j’;fSld; njh;t[ ml;il(Exam Pad) fl;lhak; bfhz;Ltut[k;/ 3/ njh;t[ miwapy; ve;jtpj kpd;dpay; kw;Wk; kpd;dDtpay; rhjd’;fis gad;gLj;jf; TlhJ/ 4/ njh;t[ vGj tUk; kDjhuh;fs; j’;fSf;F mDg;gg;gl;l mwptpg;g[ rPl;il cld; vLj;J tut[k;/ 5/ tpz;zg;gjhuh;fs;; njh;tpid ePyk;-fUik (Blue or Black Point Pen) epw ik bfhz;l vGJnfhiy gad;gLj;JkhW mwpt[Wj;jg;gLfpwJ/ 6/ kDjhuh;fSf;F j’;fspd; njh;t[ miw kw;Wk; njh;t[ neuk; ,d;Dk; rpy jpd’;fspy; http://districts.ecourts.gov.in/vellore vd;w ,izajsj;jpy; bjhptpf;fg;gLk;/ njh;t[ vGj tUk; Kd;dnu midj;J tptu’;fisa[k; mwpe;J tu ntz;Lk;/ 7/ fhyjhkjkhf tUk; ve;j kDjhuUk; njh;t[ vGj mDkjpf;fg;glkhl;lhJ/ 8/ njh;t[ vGJk; ve;j xU tpz;zg;gjhuUk; kw;wth; tpilj;jhis ghh;j;J vGjf; TlhJ. -

Higher Secondary Examination March 2020 First Year CE Not Uploaded List As on 09/06/2020
Ex-II/CEMISS-1/I/072020 Office of the Directorate of General Education Housing Board Building Shanthinagar Thiruvananthapuram 09/06/2020 Even though Missing Second Year Missing CE Mark list was published on 31/03/2020, still following CE Marks of the First year candidates are not received yet. As it will adversly affect the publication of results, all Principals are requested to verify the following list and immediately upload the CE Marks of respective candidates through IExam before 11/06/2020 2 PM. If the candidate has no CE or is not registered for the First Year March 2020 examination please specify that and send mail to "[email protected]". The results of candidates whose CE marks not received will be withheld and the Principal shall be responsible for the non publication of their results. Sd/- Joint Director Examination Higher Secondary Examination March 2020 First Year CE Not Uploaded List as on 09/06/2020 Reg No Subject School : 01020 GOVT. HSS, THOLIKODE, TRIVANDRUM 8005396 JOSBIN MOHAN English 8005396 JOSBIN MOHAN Malayalam 8005396 JOSBIN MOHAN History 8005396 JOSBIN MOHAN Economics 8005396 JOSBIN MOHAN Political Science 8005396 JOSBIN MOHAN Sociology School : 01036 GOVT GIRLS HSS, KARAMANA, TRIVANDRUM 8010134 JASNA JALEEL English 8010134 JASNA JALEEL Malayalam 8010134 JASNA JALEEL History 8010134 JASNA JALEEL Economics 8010134 JASNA JALEEL Geography 8010134 JASNA JALEEL Malayalam (Part 3) 8010135 JOBITHA B English 8010135 JOBITHA B Malayalam 8010135 JOBITHA B History 8010135 JOBITHA B Economics 8010135 JOBITHA B Geography 8010135 JOBITHA B Malayalam (Part 3) 8010143 NANDHANA K S English 8010143 NANDHANA K S Malayalam 8010143 NANDHANA K S History 8010143 NANDHANA K S Economics 8010143 NANDHANA K S Geography 8010143 NANDHANA K S Malayalam (Part 3) School : 01049 LEO XIII HSS, PULLUVILA, TRIVANDRUM 8012781 ABHIJITH. -

COMMON ELIGIBILITY TEST 2020 (For M
COMMON ELIGIBILITY TEST 2020 (For M. Phil. And Ph.D. Programs 2020-2021) Eligible Applicant List (Updated) S. No Application No Appearing Subject Name Father/Guardian Name Email Id 1 CET/2020-21/00001 Commerce DEVICHANDRIKA S B.SIVASUBRAMANIAN [email protected] 2 CET/2020-21/00002 English SINDUJA B BUPATHY G [email protected] 3 CET/2020-21/00003 Computer Science Vishali priya. O T.OOMATHURAI [email protected] 4 CET/2020-21/00004 Journalism And Mass Comm. Melwin Samuel Rajamani [email protected] 5 CET/2020-21/00005 Computer Applications Megala V Vanaja [email protected] 6 CET/2020-21/00006 Commerce PRAKASH D K R DHANABALAN [email protected] 7 CET/2020-21/00007 English PADHMAVATHI Alagarsamy ALAGARSAMY [email protected] 8 CET/2020-21/00008 Costume Design & Fashion K.SATHYA SATHYA M KRISHNASAMY [email protected] 9 CET/2020-21/00009 Mathematics GIFTEENA HINGIS Y M YESUDHASAN C [email protected] 10 CET/2020-21/00010 Chemistry SWATHI THARANI.D Dharmalingam.R [email protected] 11 CET/2020-21/00011 Computer Applications Nandhakumar SubramaniamSubbulakshmi [email protected] 12 CET/2020-21/00012 English Priyadharsini R D Rajagopal [email protected] 13 CET/2020-21/00013 Psychology KANCHANA D M DHANALAKSHMI M [email protected] 14 CET/2020-21/00014 Computer Science JINCY JOY JOY SAMUEL [email protected] 15 CET/2020-21/00015 English B. Dheepthi P C Balamohan [email protected] 16 CET/2020-21/00016 Education Dany K J Jose [email protected] 17 CET/2020-21/00017 -
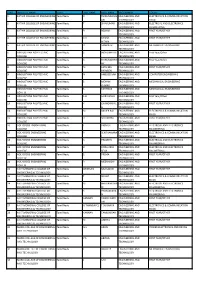
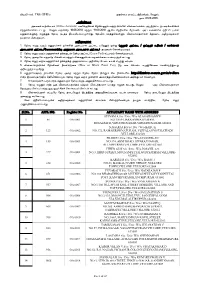
S.No Institute Name State Last Name First Name Programme
S.NO INSTITUTE NAME STATE LAST NAME FIRST NAME PROGRAMME COURSE 1 KATHIR COLLEGE OF ENGINEERING Tamil Nadu R THIRUMURUG ENGINEERING AND ELECTRONICS & COMMUNICATION AN TECHNOLOGY ENGG 2 KATHIR COLLEGE OF ENGINEERING Tamil Nadu T SIVAKUMAR ENGINEERING AND ELECTRICAL AND ELECTRONICS TECHNOLOGY ENGINEERING 3 KATHIR COLLEGE OF ENGINEERING Tamil Nadu R RESHMI ENGINEERING AND FIRST YEAR/OTHER TECHNOLOGY 4 KATHIR COLLEGE OF ENGINEERING Tamil Nadu K.V KANNA ENGINEERING AND FIRST YEAR/OTHER NITHIN TECHNOLOGY 5 KATHIR COLLEGE OF ENGINEERING Tamil Nadu R SAMPATH ENGINEERING AND MECHANICAL ENGINEERING TECHNOLOGY 6 HINDUSTHAN POLYTECHNIC Tamil Nadu S INDHUMATHI ENGINEERING AND First Year/Other COLLEGE TECHNOLOGY 7 HINDUSTHAN POLYTECHNIC Tamil Nadu K THIRUMOORT ENGINEERING AND First Year/Other COLLEGE HY TECHNOLOGY 8 HINDUSTHAN POLYTECHNIC Tamil Nadu M FATHIMA ENGINEERING AND FIRST YEAR/OTHER COLLEGE PARVEEN TECHNOLOGY 9 HINDUSTHAN POLYTECHNIC Tamil Nadu N ANBUSELVAN ENGINEERING AND COMPUTER ENGINEERING COLLEGE TECHNOLOGY 10 HINDUSTHAN POLYTECHNIC Tamil Nadu S MOHAN ENGINEERING AND MECHANICAL ENGINEERING COLLEGE KUMAR TECHNOLOGY 11 HINDUSTHAN POLYTECHNIC Tamil Nadu S KARTHICK ENGINEERING AND MECHANICAL ENGINEERING COLLEGE TECHNOLOGY 12 HINDUSTHAN POLYTECHNIC Tamil Nadu S.H SAIRA BANU ENGINEERING AND First Year/Other COLLEGE TECHNOLOGY 13 HINDUSTHAN POLYTECHNIC Tamil Nadu K CHANDRAKAL ENGINEERING AND FIRST YEAR/OTHER COLLEGE A TECHNOLOGY 14 HINDUSTHAN POLYTECHNIC Tamil Nadu M XAVIER RAJ ENGINEERING AND ELECTRONICS & COMMUNICATION COLLEGE TECHNOLOGY ENGG 15 HINDUSTHAN POLYTECHNIC Tamil Nadu R SRI VIDHYA ENGINEERING AND FIRST YEAR/OTHER COLLEGE TECHNOLOGY 16 HOLYCROSS ENGINEERING Tamil Nadu J. VIGNESH ENGINEERING AND ELECTRICAL AND ELECTRONICS COLLEGE TECHNOLOGY ENGINEERING 17 HOLYCROSS ENGINEERING Tamil Nadu P. SENTHAMARA ENGINEERING AND ELECTRONICS & COMMUNICATION COLLEGE I TECHNOLOGY ENGG 18 HOLYCROSS ENGINEERING Tamil Nadu K. -
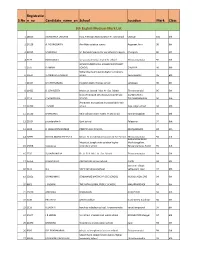
Mark List.Xlsx
Registratio S.No n_no Candidate_name_enSchool Location Mark Class 6th English Medium Mark List 1 18025 AISHWARYA LAKSHMI Indo American Matriculation Hr. Sec school Cheyyar 100 6th 2 24128 A. POONGGAVIN Aim Matriculation school Aagaram, Arni 98 6th 3 21945 S.MOKESH Sri Ramakrishna mat hr sec school chengam Chengam 96 6th 4 5177 RAKSHANA S sri saraswathi vikas matric hr.school Thiruvannamalai 96 6th GOVERNMENT GIRLS HIGHER SECONDARY 5 1575 R AMRIN SCHOOL CHEYYAR 96 6th Sidhardha matriculation higher secondary 6 20514 V.AMIRTHA VARSHINI school Peranamallur 96 6th 7 11354 M.DHEENABANU Kingston matric he.Dec.school vandavasi 96 6th 8 11422 D. JOY BLESSY Mount st. Joseph. Mat. Hr. Sec. School Tiruvannamalai 96 6th GANDHI NAGAR MATRICULATION HR SEC KIZHNACHIPET 9 3214 C M MOSHIHA SCHOOL TIRUVANNAMALAI 96 6th Amravathi murugaiyan municipal girls high 10 11700 T.SALINI school Raja ranjan street 94 6th 11 13130 DHARSAN S Vela vidhyasharam matric hr sec school Kannamangalam 94 6th 12 15334 priyadarshini k Govt school Palayanur 94 6th 13 4228 R. JASHVANTH KUMAR PINKZ PUBLIC SCHOOL KASTHAMBADI 94 6th 14 14991 NICOLA MARIA GREENE.N Mount.St.Joseph.Matriculation.Hr.Sec.School Thiruvannamalai 94 6th Avalurpettai Road, Mount.st.Joseph.matriculation higher Mathalangulam, 15 13858 S.Anusuya secondary school Tiruvannamalai, Tamil 94 6th 16 2535 SUHAIBU NISHA Dr. V. G. N. Mat. Hr. Sec. School Thiruvannamalai 94 6th 17 16354 R.SHIVARAM. AIM MATRIC Hr.Sec.School. ARANI 94 6th keeranur village, 18 9111 G.S SKV International school vettavalam road 94 6th 19 32051 SRIKRISHNA S SENDHAMIZ MATRIC HR SEC SCHOOL KILKODUNGALORE 94 6th 20 8861 J.YAZHINI THE PATH GLOBAL PUBLIC SCHOOL MALAPPAMBADI 94 6th 21 15230 ANUSHKA BHAGAVAN KILNATHUR 94 6th 22 3236 PRATAP.R GHSS Korakkai Road street, Korakkai 94 6th 23 1116 V.SACHIN kendriya vidyalaya school, Tiruvannamalai kanathampoondi 94 6th 24 14347 J R SANJAI Virutcham international public school ARUGAVOOR,cheyyar 94 6th 25 21090 MONISHA St.Antony's matriculation school Kanji 94 6th 26 9920 R.P. -

OVERALL MERIT LIST - MD MS COURSE - JULY 2019 SESSION RANK SL ROLL DOB CORRECT NEGATIVE TOTAL NAME of the CANDIDATE Percentile Eligibility NO NO
OVERALL MERIT LIST - MD MS COURSE - JULY 2019 SESSION RANK SL ROLL DOB CORRECT NEGATIVE TOTAL NAME OF THE CANDIDATE Percentile Eligibility NO NO. (dd/mm/yyyy) RESPONSE RESPONSE MARKS UR OBC SC ST INST OPH Sponsored Foreign 1 140200 PRAVEEN KUMAR 04/10/1994 134 92 444 99.988 1 1 Yes 2 110093 PATEL DARSHANKUMAR JASHVANTLAL 04/11/1995 136 104 440 99.977 2 Yes 3 201098 VOJJALA NIKHIL 22/09/1995 134 98 438 99.965 3 Yes 4 142293 MOHAMED ILIYAS R 18/03/1992 128 82 430 99.953 4 2 Yes 5 120368 BHAVESH M 29/10/1995 135 115 425 99.942 5 Yes 6 120311 SHIVAM GOEL 01/12/1994 122 74 414 99.918 6 1 Yes 7 171739 MANAV M S 15/08/1994 130 106 414 99.918 7 3 Yes 8 170591 HEMANTH AMARDEEP SANTHOOR 12/06/1995 131 115 409 99.907 8 Yes 9 170005 KUNAL SHARMA 29/04/1995 128 109 403 99.895 9 Yes 10 200687 VARIKUTI SRINADH REDDY 24/04/1995 124 94 402 99.883 10 Yes 11 171202 GAURAV SARNAIK 02/11/1995 123 92 400 99.86 11 Yes 12 160016 EVANGELINE MARY KIRUBA SAMUEL 18/05/1996 128 112 400 99.86 12 2 Yes 13 140080 J SUDHARSHAN 12/01/1996 126 108 396 99.848 13 3 Yes 14 181149 B. MITHIN KUMAR 13/12/1995 127 114 394 99.836 14 4 Yes 15 150804 SHIVAM AGARWAL 12/09/1995 125 112 388 99.825 15 Yes 16 170442 MOHIT GANDHI 23/12/1994 120 94 386 99.813 16 Yes 17 180382 JAYARAM. -

Merit List 2017-18 First Year.Pdf
Category wise Provisional Merit List of eligible students subject to verification of documents for the Grant of Pragati & Saksham (Degree & Diploma) 2017-18 (1 st year) Pragati (Degree)-Nos. of Schlarships-2000 Sl. No. Categories Merit No. Number of Student 1. Open Category 0001 to 1010 1010 2. OBC Category 1011 to 1933 540 3. SC category 1011 to 4619 300 4. ST Category 1011 to 5320 85 Pragati (Degree) 2017-18 1st year S. No. Category Merit No. Student Unique Student Name Father Name Course Name AICTE Institute InstituteName Institute District lnstitute State Id Permanent ID 1 OPEN 1 2017079460 Prateeka Bhat Chandrashekar V Bhat INFORMATION SCIENCE AND 1-5884543 B.M.S.COLLEGE OF BANGALORE URBAN Karnataka ENGINEERING ENGINEERING 2 OPEN 2 2017071837 Liliya Mary Sunny Sunny Varghese ELECTRICAL AND ELECTRONICS 1-13392996 GOVERNMENTENGIN THRISSUR Kerala ENGINEERING EERINGCOLLEGETH RISSUR 3 OPEN 3 2017071711 Jayasri Veeravilli Suryanarayana Veeravilli ELECTRONICS & COMMUNICATION 1-5788131 VALLIAMMAI KANCHIPURAM Tamil Nadu ENGG ENGINEERING COLLEGE 4 OPEN 4 2017072605 Nilla Prameela Nilla Vasu COMPUTER SCIENCE AND 1-5906491 SAGI WEST GODAVARI Andhra Pradesh ENGINEERING RAMAKRISHNAM RAJU ENGINEERING COLLEGE 5 OPEN 5 2017083736 Kamani Ramya Kamani Ravinder COMPUTER SCIENCE & ENGINEERING 1-495566375 JNTUH COLLEGE OF RANGAREDDI Telangana ENGINEERING HYDERABAD 6 OPEN 6 2017078875 Elagandula Nikhitha Elagandula Ashok CIVIL ENGINEERING 1-495566375 JNTUH COLLEGE OF RANGAREDDI Telangana ENGINEERING HYDERABAD 7 OPEN 7 2017085401 Susmitha Naga Lakshmi Narayana -

Dr. NTR UNIVERSITY of HEALTH SCIENCES :: A.P :: VIJAYAWADA – 08 MBBS,BDS ADMISSIONS 2020-21 UNDER MANAGEMENT QUOTA ( Category
Dr. NTR UNIVERSITY OF HEALTH SCIENCES :: A.P :: VIJAYAWADA – 08 MBBS,BDS ADMISSIONS 2020-21 UNDER MANAGEMENT QUOTA ( Category - B & C (NRI) ) PROVISIONAL MERIT POSITION OF CANDIDATES APPLIED -AFTER VERIFICATION OF UPLOADED CERTIFICATES Note: 1. Candidates are informed to submit their grievances, if any, in the displayed provisional merit position, with regard to his/her details ( Gender, NRI & Minority) , it should be brought to the notice immediately through the e-mail to [email protected] with proper supporting documents to reach by 3.00 PM on 19-12-2020 duly mentioning their Rank , Roll Number 2. The Grievance received after the above said time and date will not be considered 3. Grievances with out proper documents will not be considered. 4. Merit lists of eligible candidates for NRI category seats in Sri Padmavathi Medical College for Women will be displayed after receiving from the SVIMS authorities. M.O RANK Roll No STUDENT_NAME SX CAT NRI MIN 1 28020 3104204066 YUSUF AHMED M OC NO MSM 2 32664 3118030130 SHRAWAN KUMAR M OC NO 3 34643 4408131052 A N SAI KARTHIK M OC NO 4 41441 3605005158 SIPRA PATTNAIK F OC NO 5 41897 1205016043 CHUNDURU LALITH DURGA PREETHAM M OC NO 6 43392 3605005256 ANANYA MAHAPRASHASTA F OC NO 7 44033 1205007152 PUPPALA LAKSHMI NAGA PRAHARSHA F OC NO 8 44631 1201002115 NUTHALAPATI GOWTHAM M OC NO 9 44910 1205010199 RAMIREDDY KRISHNAA TEJESWAAR REDDY M OC NO 10 45128 3903120171 RAMESH DEWASI M OBC NO 11 45939 1205011329 KONETI RITHIN KAMALAKAR M OC NO 12 46656 1201015005 POOJITHA AAKI F OC NO 13 46658 -

Tamil Nadu Public Service Commission Bulletin Extraordinary
© [Regd. No. TN/CCN-466/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 196/2009 2018 [Price: Rs. 146.40 Paise. TAMIL NADU PUBLIC SERVICE COMMISSION BULLETIN EXTRAORDINARY No. 6] CHENNAI, WEDNESDAY, MARCH 7, 2018 Maasi 23, Hevilambi, Thiruvalluvar Aandu-2049 CONTENTS DEPARTMENTAL TESTS—RESULTS, DECEMBER 2017 NAME OF THE TESTS AND CODE NUMBERS PPagesages PPagesages Subordinate Accounts Services Examinations Tamil Nadu Wakf Board Department Test - First Paper - Iii (Accounts and Audit in internal Paper (Detailed Application) (Without Books Audit and Statutory Boards Audit) (Without & With Books) (Test Code No. 171) .. 1193-19493-194 Books & With Books) (Test Code No. 138) 187-189 Panchayat Development Account Test (Without Departmental Test for Audit Superintendents Books & With Books) (Test Code No. 166) 1194-20494-204 of Highways Department - Second Paper (theoretical and Practical (Without Books the Account Test for Subordinate offi cers - & With Books) (Test Code No. 139) .. Part - I (With Out Books & With Books) .. 118989 (Test Code No. 124) .. .. .. .. 2205-28105-281 Departmental Test for Audit Superintendents Advanced Language Test for officers of of Highways Department - Fifth Paper the Tamil Nadu Educational Subordinate (Establishment Audit - theoritical and Service - Tamil First Paper (Prose and Practical) (Without Books & With Books) Poetry) (Without Books) (Test Code No. (Test Code No. 140) .. .. .. .. 1189-19089-190 034) .. 2281-28381-283 Departmental Test for Local Fund Audit and Advanced Language Test for officers of internal Audit Departments (Without Books the Tamil Nadu Educational Subordinate & With Books) (Test Code No. 153) .. 119090 Service - Tamil Second Paper (Translation and Essay) (Without Books) (Test Code Tamil Nadu Water Supply and Drainage Board No. -

In the High Court of Karnataka, Bengaluru
1 R IN THE HIGH COURT OF KARNATAKA, BENGALURU DATED THIS THE 21 st DAY OF DECEMBER, 2018 BEFORE THE HON'BLE MR. JUSTICE KRISHNA S.DIXIT WRIT PETITION NOs. 46917-47025 OF 2018 (EDN-RES) C/W WRIT PETITION NOS. 45738 OF 2018, 46335- 46523/2018(EDN-RES), 47256-47276 OF 2018 (S-RES), 52140-52172/2018(EDN-RES) IN W.P. NOs. 46917-47025 OF 2018: BETWEEN: 1. DR SWAMY MANJUNATH S T S/O MANJUNATH S AGED ABOUT 28 YEARS RESIDING AT MAHADEVI NILAYA OPPOSITE KEB OFFICE JYOTHI NAGARA, SIRA, TUMKUR - 572137 2. DR SAMEERA D/O IBRAHEEM PATEL AGED ABOUT 28 YEARS RESIDING AT # 7-844/B MEHBOOB MANZIL MIJGORI NAYA MOHALLA GULBARGA - 585104 3. DR ASHWINI B S D/O DR B N SHIVARAM AGED ABOUT 29 YEARS RESIDING AT NO H NO 28 SAI NILAYA, NEAR PADMA CHITHRAMANDIRA AMRUTHAHALLI, BANGALORE - 92 4. DR ANUSHA K L D/O LAKSHMANA AGED ABOUT 28 YEARS RESIDING AT 11, SRI GURU APRAMEYA SADANA 3RD MAINROAD, ANANTHAPURA YELAHANKA, BENGALURU - 560064 2 5. DR SOWMY A J D/O JAGADEESHWARA V AGED ABOUT 28 YEARS RESIDING AT G5, SILVER WAVE ELEGANCE BEHIND BETHESDA CHURCH VALAGEREHALLI KENGERI SATELLITE TOWN BANGALORE - 560060 6. DR SHALOM ELSY PHILIP D/O SAJI PHILIP AGED ABOUT 27 YEARS RESIDING AT TALENTS FOR CHIRST PALLICKAL P O KATTANAM - 690503 7. DR SHETTY HARSHAVARADHAN KUSHAL S/O KUSHAL SHETTY AGED ABOUT 29 YEARS RESIDING AT 14-145/A 3, AIRPORT ROAD, 403, ADITYA COMPLEX, NEAR MASHAAL CHOWK NANI DAMAN, DAMAN U T - 396210 8. DR SUPRITHA J C D/O CHANNAPPA J B AGED ABOUT 27 YEARS RESIDING AT NO 1308 SPANDANA NILAYA, 3 RD CROSS, NEHRU NAGAR, MANDYA - 571401 9. -
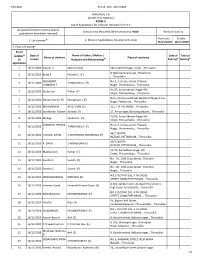
ANNEXURE 5.8 (CHAPTER V, PARA 25) FORM 9 List of Applica Ons For
11/23/2020 Form9_AC6_23/11/2020 ANNEXURE 5.8 (CHAPTER V, PARA 25) FORM 9 List of Applicaons for inclusion received in Form 6 Designated locaon identy (where Constuency (Assembly/£Parliamentary): Avadi Revision identy applicaons have been received) From date To date @ 2. Period of applicaons (covered in this list) 1. List number 16/11/2020 16/11/2020 3. Place of hearing* Serial number$ Date of Name of Father / Mother / Date of Time of Name of claimant Place of residence of receipt Husband and (Relaonship)# hearing* hearing* applicaon 1 16/11/2020 Naresh S Meena S (M) 540, Lakshmi Nagar, Avadi, , Thiruvallur 8, Bakkiyammal street , Paabiram 2 16/11/2020 Balaji P Prakash S (F) , , Thiruvallur YASHWANT No.3, 3 rd cross street, Prakash 3 16/11/2020 THANGAVEL N (F) KUMAR N T Nagar, Thiruninravur, , Thiruvallur 91/65, Annai Abirami Nagar 5th 4 16/11/2020 Sasikumar Pithan (F) Street, Thiruverkadu, , Thiruvallur No 5, Immanuvel Street,Senthamil Nagar,Anna 5 16/11/2020 Kishore Kumar M Murugesan C (F) Nagar, Paabiram, , Thiruvallur 6 16/11/2020 MOHANRAJ A ACHUTHAN (F) 215, T N H B, AVADI, , Thiruvallur 7 16/11/2020 Sasidharan Rasaiah Rasaiah (F) 17, Anna nagar, Kannadapalayam, , Thiruvallur 91/65, Annai Abirami Nagar 5th 8 16/11/2020 Malliga Sasikumar (H) Street, Thiruverkadu, , Thiruvallur SANDHIYA PRIYA N No.3, 3 rd cross street, Prakash 9 16/11/2020 THANGAVEL N (F) T Nagar, Thiruninravur, , Thiruvallur 46/7, NORTH 10 16/11/2020 A DAVID DAVID S ABISHEGAM ABISHEGAM (F) BAZAAR, PATTABIRAM, , Thiruvallur 46/7, NORTH 11 16/11/2020 A DAVID S ABISHEGAM -

Tamil Nadu Government Gazette
© [Regd. No. TN/CCN/467/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 197/2009. 2019 [Price : Rs. 315.20 Paise. TAMIL NADU GOVERNMENT GAZETTE PUBLISHED BY AUTHORITY No. 25A] CHENNAI, WEDNESDAY, JUNE 19, 2019 Aani 4, Vikari, Thiruvalluvar Aandu–2050 Part VI–Section 1 (Supplement) NOTIFICations BY HEADS OF departments, ETC. TAMIL NADU MEDICAL COUNCIL Supplementary Medical Register for the year 2018 DTP—VI-1-Sup. (25A)—1 [ 1 ] 12 REGISTRATION FEES Rs. P. (a) For First Registration (other than reciprocal registration) under section 14 of 3,500.00/- the Tamil Nadu Medical Registration Act,1914, including Stamp Fees Act postage for a free copy of the Tamil Nadu Medical Register, Smart Name Board and Identity card (b) For Foreign Medical Graduates Permanent Registration 10,000.00/- including Stamp Fees Act postage for a free copy of the Tamil Nadu Medical Register, Smart Name Board and Identity card (c) For the Registration of each Additional Qualification obtained –Degree 4,000-00/- subsequent to first registration including Smart Name Board Diploma 3,000-00 /- and Identity Card Superspeciality 5,000-00/- (d) For Change of Name 2,000.00 (e) For Reciprocal Registration Nil (f) For Duplicate Copy of Registration Certificate including stamp fee 2,000.00/- (g)For Provisional Registration 1,500.00/- (h)For Foreign Medical Graduates Provisional Registration Certificate 5,000.00/- (i) For Duplicate Copy of Provisional Registration Certificate 1,000.00/- (j) For Tamil Nadu Medical Council Certificate of Good standing 5,000.00/- (k) For No Objection Certificate 2,000.00/- (l) For C.T.Forms ,E.C.M.G.(Verification) 5,000.00/- (m)For New Medical Registration Certificate including stamp fee 1,000.00/- (n) For Smart Name Board 800.00/- (o) For Identity Card 150.00/- (p) Other Miscellaneous Items Endorsement in Certificates (Date Extension & Name & Initial Correction) and Registration related Matters.