High-Performance Web Services for Querying Gene and Variant Annotation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Interaction Between Web Browsers and Script Engines
IT 12 058 Examensarbete 45 hp November 2012 Interaction between web browsers and script engines Xiaoyu Zhuang Institutionen för informationsteknologi Department of Information Technology Abstract Interaction between web browser and the script engine Xiaoyu Zhuang Teknisk- naturvetenskaplig fakultet UTH-enheten Web browser plays an important part of internet experience and JavaScript is the most popular programming language as a client side script to build an active and Besöksadress: advance end user experience. The script engine which executes JavaScript needs to Ångströmlaboratoriet Lägerhyddsvägen 1 interact with web browser to get access to its DOM elements and other host objects. Hus 4, Plan 0 Browser from host side needs to initialize the script engine and dispatch script source code to the engine side. Postadress: This thesis studies the interaction between the script engine and its host browser. Box 536 751 21 Uppsala The shell where the engine address to make calls towards outside is called hosting layer. This report mainly discussed what operations could appear in this layer and Telefon: designed testing cases to validate if the browser is robust and reliable regarding 018 – 471 30 03 hosting operations. Telefax: 018 – 471 30 00 Hemsida: http://www.teknat.uu.se/student Handledare: Elena Boris Ämnesgranskare: Justin Pearson Examinator: Lisa Kaati IT 12 058 Tryckt av: Reprocentralen ITC Contents 1. Introduction................................................................................................................................ -

The NIDDK Information Network: a Community Portal for Finding Data, Materials, and Tools for Researchers Studying Diabetes, Digestive, and Kidney Diseases
RESEARCH ARTICLE The NIDDK Information Network: A Community Portal for Finding Data, Materials, and Tools for Researchers Studying Diabetes, Digestive, and Kidney Diseases Patricia L. Whetzel1, Jeffrey S. Grethe1, Davis E. Banks1, Maryann E. Martone1,2* 1 Center for Research in Biological Systems, University of California, San Diego, San Diego, California, a11111 United States of America, 2 Dept of Neurosciences, University of California, San Diego, San Diego, California, United States of America * [email protected] Abstract OPEN ACCESS The NIDDK Information Network (dkNET; http://dknet.org) was launched to serve the needs Citation: Whetzel PL, Grethe JS, Banks DE, Martone of basic and clinical investigators in metabolic, digestive and kidney disease by facilitating ME (2015) The NIDDK Information Network: A access to research resources that advance the mission of the National Institute of Diabe- Community Portal for Finding Data, Materials, and Tools for Researchers Studying Diabetes, Digestive, tes and Digestive and Kidney Diseases (NIDDK). By research resources, we mean the and Kidney Diseases. PLoS ONE 10(9): e0136206. multitude of data, software tools, materials, services, projects and organizations available to doi:10.1371/journal.pone.0136206 researchers in the public domain. Most of these are accessed via web-accessible data- Editor: Chun-Hsi Huang, University of Connecticut, bases or web portals, each developed, designed and maintained by numerous different UNITED STATES projects, organizations and individuals. While many of the large government funded data- Received: February 17, 2015 bases, maintained by agencies such as European Bioinformatics Institute and the National Accepted: July 30, 2015 Center for Biotechnology Information, are well known to researchers, many more that have been developed by and for the biomedical research community are unknown or underuti- Published: September 22, 2015 lized. -

High-Performance Web Services for Querying Gene
Xin et al. Genome Biology (2016) 17:91 DOI 10.1186/s13059-016-0953-9 SOFTWARE Open Access High-performance web services for querying gene and variant annotation Jiwen Xin1†, Adam Mark1,2†, Cyrus Afrasiabi1†, Ginger Tsueng1, Moritz Juchler3, Nikhil Gopal3, Gregory S. Stupp1, Timothy E. Putman1, Benjamin J. Ainscough4, Obi L. Griffith4, Ali Torkamani5,6, Patricia L. Whetzel7, Christopher J. Mungall8, Sean D. Mooney3, Andrew I. Su1,6* and Chunlei Wu1* Abstract Efficient tools for data management and integration are essential for many aspects of high-throughput biology. In particular, annotations of genes and human genetic variants are commonly used but highly fragmented across many resources. Here, we describe MyGene.info and MyVariant.info, high-performance web services for querying gene and variant annotation information. These web services are currently accessed more than three million times per month. They also demonstrate a generalizable cloud-based model for organizing and querying biological annotation information. MyGene.info and MyVariant.info are provided as high-performance web services, accessible at http://mygene.info and http://myvariant.info. Both are offered free of charge to the research community. Keywords: Annotation, Gene, Variant, API, Cloud, Repository, Database Background services. Federated data solutions are always up to date, The accumulation of biomedical knowledge is growing but extra care is required to maintain the links, and exponentially. There has been tremendous effort to large queries may take a long time to return due to ser- structure research findings as annotations on biological ver and network limitations. Moreover, the dependability entities (e.g., genes, genetic variants, and pathways). -

Audience-Driven Web Design
Updated version published in: Information modelling in the new millennium, Eds. Matt Rossi & Keng Siau, IDEA GroupPublishing, ISBN 1-878289-77-2, 2001. Audience-driven Web Design Olga De Troyer Vrije Universiteit Brussel WISE Research group Pleinlaan 2, B-1050 Brussel, Belgium [email protected] INTRODUCTION Today Web-related software development seems to be faced with a crisis not unlike the one that occurred a generation ago when in the 70s computer hardware experienced an order of magnitude increase in computational power. While this made possible the implementation of a new class of applications larger both in size and complexity, the methods for software development available at that time were not able to scale up to such large projects. The "software crisis" was a fact with its legendary stories of delays, unreliability, maintenance bottlenecks and costs. Now we seem to be starting to deal painfully with a corresponding "web site crisis". Over the last few years, the Internet has boomed and the World Wide Web with it. Web browsers are the basic user platform of the Internet. Because of the immense potential audience, and because publishing on the web is in principle very easy, the number of web applications has exploded. Most of the web sites are created opportunistically without prior planning or analysis. Moreover, even large mission-critical intranet projects are being started without any regard for methodology. The resulting problems of maintenance and development backlog, so well known in "classical" information systems, can easily be predicted, and will happen on a much larger scale. Because web sites are almost by definition required to adapt and grow, and have to interact with other sites and systems unknown at the moment of creation, these problems will also be much more complex and severe. -
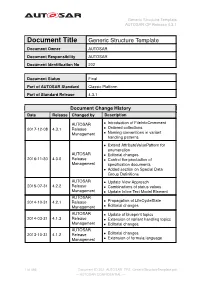
Generic Structure Template AUTOSAR CP Release 4.3.1
Generic Structure Template AUTOSAR CP Release 4.3.1 Document Title Generic Structure Template Document Owner AUTOSAR Document Responsibility AUTOSAR Document Identification No 202 Document Status Final Part of AUTOSAR Standard Classic Platform Part of Standard Release 4.3.1 Document Change History Date Release Changed by Description AUTOSAR • Introduction of FileInfoCmmment 2017-12-08 4.3.1 Release • Ordered collections Management • Naming conventions in variant handling patterns • Extend AttributeValuePattern for enumeration AUTOSAR • Editorial changes 2016-11-30 4.3.0 Release • Control the production of Management specification documents • Added section on Special Data Group Definitions AUTOSAR • Update View Approach 2015-07-31 4.2.2 Release • Combinations of status values Management • Update Inline Text Model Element AUTOSAR 2014-10-31 4.2.1 Release • Propagation of LifeCycleState Management • Editorial changes AUTOSAR • Update of blueprint topics 2014-03-31 4.1.3 Release • Extension of variant handling topics Management • Editorial changes AUTOSAR 2013-10-31 4.1.2 Release • Editorial changes Management • Extension of formula language 1 of 456 Document ID 202: AUTOSAR_TPS_GenericStructureTemplate.pdf — AUTOSAR CONFIDENTIAL — Generic Structure Template AUTOSAR CP Release 4.3.1 • Editorial changes including tagged specification items AUTOSAR • Support of build action manifest 2013-03-15 4.1.1 Administration • Support of roles and rights • Added life cycle support • Support of collections and collectable elements • Editorial changes including -

Use Cases and Software Source Code Identification SCID WG
https://tinyurl.com/qpg7n7m Use Cases and Software Source Code Identification SCID WG 27th March 2020 - RDA 15th Plenary Virtual Meeting https://tinyurl.com/qpg7n7m Contributors (alphabetical order by name) ● Alice Allen, Astronomy Source Code Library & U. Maryland, USA ● Anita Bandrowski - University of California San Diego, USA ● Roberto Di Cosmo - Software Heritage, Inria and University of Paris, France ● Martin Fenner - DataCite, Germany ● Morane Gruenpeter - Inria, Software Heritage, France ● Daniel S. Katz - University of Illinois at Urbana-Champaign, USA ● John Kunze - California Digital Library, University of California, USA ● Moritz Schubotz - swMATH, FIZ Karlsruhe, Germany Agenda https://tinyurl.com/qpg7n7m ● Introduction ○ Objectives, Outputs, Connections to other WG ● Use cases (Archiving, Referencing, Describing, Citing, etc.) ○ Audience may suggest additional use cases ● Identifiers schemas ○ DOIs, Hashes, SWH-ID,Wikidata entities, ARKS, ASCL-ID, RRID, swMATH-ID ● Small group discussion ○ documenting the use case : challenges, pros, and cons of different identifiers per use case ● Report-back and discussion ● Wrap-up https://tinyurl.com/qpg7n7m Introduction Software Source Code Identification Working Group Chronology... Co-chairs Spawned at RDA P11 in Berlin from ● Roberto Di Cosmo ● the RDA SSC IG & ● Martin Fenner ● the FORCE11 SCIWG ● Daniel S. Katz 10/2018 - TAB endorsement Web page https://www.rd-alliance.org/groups/software-source-code-iden tification-wg 4/2019 - RDA P13, Philadelphia ● WG kick-off Repository https://github.com/force11/force11-rda-scidwg -

International Workshop on Software Variability Management
International Workshop on Software Variability Management (SVM) ICSE’03 International Conference on Software Engineering Portland, Oregon May 3-11, 2003 Table Of Contents Software Configuration Management Problems and Solutions to Software Variability Management Lars Bendix, Lund Institute of Technology. 5 Using UML Notation Extensions to Model Variability in Product-line Architectures Liliana Dobrica, University Politehnica of Bucharest; Eila Niemelä, VTT Electronics. 8 Consolidating Variability Models Birgit Geppert, Frank Roessler, David M. Weiss, Avaya Labs Research . 14 Managing Variability with Configuration Techniques Andreas Hein, John MacGregor, Robert Bosch Corporation . 19 Supporting the Product Derivation Process with a Knowledge-based Approach Lothar Hotz, Thorsten Krebs, Universität Hamburg . 24 Variability Analysis for Communications Software Chung- Horng Lung, Carleton University. 30 Evolving Quality Attribute Variability Amar Ramdane-Cherif, Samir Benarif, Nicole Levy, PRISM, Université de Versailles St.- Quentin; F. Losavio, Universidad Central de Venezuela. 33 A Practical Approach To Full-Life Cycle Variability Management Klaus Schmid, Isabel John, Fraunhofer Institute for Experimental Software Engineering (IESE) . 41 Capturing Timeline Variability with Transparent Configuration Environments Eelco Dolstra, Utrecht University; Gert Florijn, SERC; Merijn de Jonge, CWI; Eelco Visser, Utrecht University . 47 Component Interactions as Variability Management Mechanisms in Product Line Architectures M. S. Rajasree, D. JanakiRam, -

(12) United States Patent (10) Patent No.: US 8,812,422 B2 Nizzari Et Al
USOO8812422B2 (12) United States Patent (10) Patent No.: US 8,812,422 B2 NiZZari et al. (45) Date of Patent: Aug. 19, 2014 (54) VARLANT DATABASE (56) References Cited (71) Applicants: Marcia M. Nizzari, Needham, MA U.S. PATENT DOCUMENTS (US); Benjamin H. Breton, Woburn, 4,683, 195 A 7, 1987 Mullis et al. MA (US); David L. Tefft, Malden, MA 4,683.202 A 7, 1987 Mullis 4,988,617 A 1/1991 Landegren et al. (US); Xavier S. Haurie, Belmont, MA 5,234,809 A 8, 1993 Boom et al. (US) 5,242,794. A 9/1993 Whiteley et al. 5,494,810 A 2/1996 Barany et al. 5,583,024 A 12/1996 McElroy (72) Inventors: Marcia M. Nizzari, Needham, MA 5,604,097 A 2f1997 Brenner (US); Benjamin H. Breton, Woburn, 5,636,400 A 6/1997 Young MA (US); David L. Tefft, Malden, MA 5,674,713 A 10/1997 McElroy (US); Xavier S. Haurie, Belmont, MA (US) (Continued) FOREIGN PATENT DOCUMENTS (73) Assignee: Good Start Genetics, Inc., Cambridge, MA (US) EP 1321477 A1 6, 2003 EP 1564306 A2 8, 2005 (*) Notice: Subject to any disclaimer, the term of this (Continued) patent is extended or adjusted under 35 OTHER PUBLICATIONS U.S.C. 154(b) by 13 days. Sequeira, R. A. R. L. Olson, and J. M. McKinion. “Implementing generic, object-oriented models in biology.” Ecological Modelling (21) Appl. No.: 13/667,575 94.1 (1997): 17-31.* (22) Filed: Nov. 2, 2012 (Continued) Primary Examiner — Kakali Chaki (65) Prior Publication Data Assistant Examiner — Kevin W Figueroa (74) Attorney, Agent, or Firm — Thomas C. -

Release 1.1.3
HGVS Release 1.1.3 Jul 02, 2018 Contents 1 Contents 3 1.1 Introduction...............................................3 1.2 Quick Start................................................5 1.3 Installing hgvs..............................................8 1.4 Key Concepts............................................... 10 1.5 Examples................................................. 13 1.6 Reference Manual............................................ 18 1.7 Privacy Issues.............................................. 49 1.8 Contributing............................................... 51 1.9 Getting Help............................................... 55 1.10 Change Log............................................... 56 1.11 License.................................................. 81 2 Indices and tables 87 Python Module Index 89 i ii HGVS, Release 1.1.3 hgvs is a Python package to parse, format, validate, normalize, and map biological sequence variants according to recommendations of the Human Genome Variation Society. Source| Documentation| Discuss| Issues Contents 1 HGVS, Release 1.1.3 2 Contents CHAPTER 1 Contents 1.1 Introduction Genome, transcript, and protein sequence variants are typically reported using the variation nomenclature (“varnomen”) recommendations provided by the Human Genome Variation Society (HGVS)(Taschner and den Dun- nen, 2011). Most variants are deceptively simple looking, such as NM_021960.4:c.740C>T. In reality, the varnomen standard provides for much more complex concepts and representations. As high-throughput sequencing -
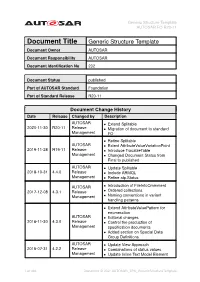
Generic Structure Template AUTOSAR FO R20-11
Generic Structure Template AUTOSAR FO R20-11 Document Title Generic Structure Template Document Owner AUTOSAR Document Responsibility AUTOSAR Document Identification No 202 Document Status published Part of AUTOSAR Standard Foundation Part of Standard Release R20-11 Document Change History Date Release Changed by Description AUTOSAR • Extend Splitable 2020-11-30 R20-11 Release • Migration of document to standard Management FO • Refine Splitable AUTOSAR • Extent AttributeValueVariationPoint 2019-11-28 R19-11 Release • Introduce TracableTable Management • Changed Document Status from Final to published AUTOSAR • Update Splitable 2018-10-31 4.4.0 Release • Include ARMQL Management • Refine atp.Status AUTOSAR • Introduction of FileInfoCmmment 2017-12-08 4.3.1 Release • Ordered collections Management • Naming conventions in variant handling patterns • Extend AttributeValuePattern for enumeration AUTOSAR • Editorial changes 2016-11-30 4.3.0 Release • Control the production of Management specification documents • Added section on Special Data Group Definitions AUTOSAR • Update View Approach 2015-07-31 4.2.2 Release • Combinations of status values Management • Update Inline Text Model Element 1 of 498 Document ID 202: AUTOSAR_TPS_GenericStructureTemplate Generic Structure Template AUTOSAR FO R20-11 AUTOSAR 2014-10-31 4.2.1 Release • Propagation of LifeCycleState Management • Editorial changes AUTOSAR • Update of blueprint topics 2014-03-31 4.1.3 Release • Extension of variant handling topics Management • Editorial changes AUTOSAR 2013-10-31 4.1.2 -

UC San Diego Previously Published Works
UC San Diego UC San Diego Previously Published Works Title The Resource Identification Initiative: A cultural shift in publishing. Permalink https://escholarship.org/uc/item/07s7d2cd Authors Bandrowski, Anita Brush, Matthew Grethe, Jeffery S et al. Publication Date 2015 DOI 10.12688/f1000research.6555.2 Peer reviewed eScholarship.org Powered by the California Digital Library University of California F1000Research 2015, 4:134 Last updated: 20 NOV 2015 RESEARCH ARTICLE The Resource Identification Initiative: A cultural shift in publishing [version 2; referees: 2 approved] Anita Bandrowski1, Matthew Brush2, Jeffery S. Grethe1, Melissa A. Haendel2, David N. Kennedy3, Sean Hill4, Patrick R. Hof5, Maryann E. Martone1, Maaike Pols6, Serena Tan7, Nicole Washington8, Elena Zudilova-Seinstra9, Nicole Vasilevsky2, Resource Identification Initiative Members are listed here: https://www.force11.org/node/4463/members 1Center for Research in Biological Systems, UCSD, la Jolla, CA, 92093, USA 2Department of Medical Informatics & Clinical Epidemiology, OHSU, Portland, Oregon, 97239, USA 3Department of Psychiatry, University of Massachusetts Medical School, Worcester, MA, 01605, USA 4Karolinska Institutet, Stockholm, 171 77, Sweden 5Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA 6Scientific Outreach, Faculty of 1000 Ltd, London, W1T 4LB, UK 7John Wiley and Sons, Hoboken, NJ, 07030, USA 8Lawrence Berkeley National Laboratory, Lawrence Berkeley National Laboratory, Berkeley, CA, 94720, USA 9Elsevier, Amsterdam, 1043 NX, Netherlands v2 First published: 29 May 2015, 4:134 (doi: 10.12688/f1000research.6555.1) Open Peer Review Latest published: 19 Nov 2015, 4:134 (doi: 10.12688/f1000research.6555.2) Referee Status: Abstract A central tenet in support of research reproducibility is the ability to uniquely identify research resources, i.e., reagents, tools, and materials that are used to Invited Referees perform experiments. -

An Analysis of the Hypertext Versioning Domain
UNIVERSITY OF CALIFORNIA, IRVINE An Analysis of the Hypertext Versioning Domain DISSERTATION submitted in partial satisfaction of the degree requirements for the degree of DOCTOR OF PHILOSOPHY in Information and Computer Science by Emmet James Whitehead, Jr. Dissertation Committee: Professor Richard N. Taylor, Chair Professor Mark S. Ackerman Professor David S. Rosenblum 2000 © 2000 Emmet James Whitehead, Jr. The dissertation of Emmet James Whitehead, Jr. is approved and is acceptable in quality and form for publication on microfilm: ___________________________________ ___________________________________ ___________________________________ Committee Chair University of California, Irvine 2000 ii Table of Contents List of Figures .............................................................................................................................................. vi List of Tables..............................................................................................................................................viii Acknowledgements ...................................................................................................................................... ix Curriculum Vitae......................................................................................................................................... xi Abstract of the Dissertation ....................................................................................................................... xv Chapter 1 – Introduction ............................................................................................................................