Alu Insertion Polymorphisms Shared by Papio Baboons and Theropithecus Gelada Reveal an Intertwined Common Ancestry Jerilyn A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Targets, Tactics, and Cooperation in the Play Fighting of Two Genera of Old World Monkeys (Mandrillus and Papio): Accounting for Similarities and Differences
2019, 32 Heather M. Hill Editor Peer-reviewed Targets, Tactics, and Cooperation in the Play Fighting of Two Genera of Old World Monkeys (Mandrillus and Papio): Accounting for Similarities and Differences Kelly L. Kraus1, Vivien C. Pellis1, and Sergio M. Pellis1 1 Department of Neuroscience, University of Lethbridge, Canada Play fighting in many species involves partners competing to bite one another while avoiding being bitten. Species can differ in the body targets that are bitten and the tactics used to attack and defend those targets. However, even closely related species that attack and defend the same body target using the same tactics can differ markedly in how much the competitiveness of such interactions is mitigated by cooperation. A degree of cooperation is necessary to ensure that some turn-taking between the roles of attacker and defender occurs, as this is critical in preventing play fighting from escalating into serious fighting. In the present study, the dyadic play fighting of captive troops of 4 closely related species of Old World monkeys, 2 each from 2 genera of Papio and Mandrillus, was analyzed. All 4 species have a comparable social organization, are large bodied with considerable sexual dimorphism, and are mostly terrestrial. In all species, the target of biting is the same – the area encompassing the upper arm, shoulder, and side of the neck – and they have the same tactics of attack and defense. However, the Papio species exhibit more cooperation in their play than do the Mandrillus species, with the former using tactics that make biting easier to attain and that facilitate close bodily contact. -

Rhesus Macaque Sequencing
White Paper for Complete Sequencing of the Rhesus Macaque (Macaca mulatta) Genome Jeffrey Rogers1, Michael Katze 2, Roger Bumgarner2, Richard A. Gibbs 3 and George M. Weinstock3 I. Introduction Humans are members of the Order Primates and our closest evolutionary relatives are other primate species. This makes primate models of human disease particularly important, as the underlying physiology and metabolism, as well as genomic structure, are more similar to humans than are other mammals. Chimpanzees (Pan troglodytes) are the animals most similar to humans in overall DNA sequence, with interspecies sequence differences of approximately 1- 1.5% (Stewart and Disotell 1998, Page and Goodman 2001). The other apes, including gorillas and orangutans are nearly as similar to humans. The animals next most closely related to humans are Old World monkeys, superfamily Cercopithecoidea. This group includes the common laboratory species of rhesus macaque (Macaca mulatta), baboon (Papio hamadryas), pig-tailed macaque (Macaca nemestrina) and African green monkey (Chlorocebus aethiops). The human evolutionary lineage separated from the ancestors of chimpanzees about 6-7 million years ago (MYA), while the human/ape lineage diverged from Old World monkeys about 25 MYA (Stewart and Disotell 1998), and from another important primate group, the New World monkeys, more than 35-40 MYA. In comparison, humans diverged from mice and other non- primate mammals about 65-85 MYA (Kumar and Hedges 1998, Eizirik et al 2001). In the evaluation of primate candidates for genome sequencing there should be more to selection of an organism than evolutionary considerations. The chimpanzee’s status as Closest Relative To Human has earned it an exemption from this consideration. -

Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea
Primate Conservation 2009 (24): 99–105 Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea Thomas M. Butynski¹,², Yvonne A. de Jong² and Gail W. Hearn¹ ¹Bioko Biodiversity Protection Program, Drexel University, Philadelphia, PA, USA ²Eastern Africa Primate Diversity and Conservation Program, Nanyuki, Kenya Abstract: Bioko Island, Equatorial Guinea, has a rich (eight genera, 11 species), unique (seven endemic subspecies), and threat- ened (five species) primate fauna, but the taxonomic status of most forms is not clear. This uncertainty is a serious problem for the setting of priorities for the conservation of Bioko’s (and the region’s) primates. Some of the questions related to the taxonomic status of Bioko’s primates can be resolved through the statistical comparison of data on their body measurements with those of their counterparts on the African mainland. Data for such comparisons are, however, lacking. This note presents the first large set of body measurement data for each of the seven species of monkeys endemic to Bioko; means, ranges, standard deviations and sample sizes for seven body measurements. These 49 data sets derive from 544 fresh adult specimens (235 adult males and 309 adult females) collected by shotgun hunters for sale in the bushmeat market in Malabo. Key Words: Bioko Island, body measurements, conservation, monkeys, morphology, taxonomy Introduction gordonorum), and surprisingly few such data exist even for some of the more widespread species (for example, Allen’s Comparing external body measurements for adult indi- swamp monkey Allenopithecus nigroviridis, northern tala- viduals from different sites has long been used as a tool for poin monkey Miopithecus ogouensis, and grivet Chlorocebus describing populations, subspecies, and species of animals aethiops). -

The Taxonomy of Primates in the Laboratory Context
P0800261_01 7/14/05 8:00 AM Page 3 C HAPTER 1 The Taxonomy of Primates T HE T in the Laboratory Context AXONOMY OF P Colin Groves RIMATES School of Archaeology and Anthropology, Australian National University, Canberra, ACT 0200, Australia 3 What are species? D Taxonomy: EFINITION OF THE The biological Organizing nature species concept Taxonomy means classifying organisms. It is nowadays commonly used as a synonym for systematics, though Disagreement as to what precisely constitutes a species P strictly speaking systematics is a much broader sphere is to be expected, given that the concept serves so many RIMATE of interest – interrelationships, and biodiversity. At the functions (Vane-Wright, 1992). We may be interested basis of taxonomy lies that much-debated concept, the in classification as such, or in the evolutionary implica- species. tions of species; in the theory of species, or in simply M ODEL Because there is so much misunderstanding about how to recognize them; or in their reproductive, phys- what a species is, it is necessary to give some space to iological, or husbandry status. discussion of the concept. The importance of what we Most non-specialists probably have some vague mean by the word “species” goes way beyond taxonomy idea that species are defined by not interbreeding with as such: it affects such diverse fields as genetics, biogeog- each other; usually, that hybrids between different species raphy, population biology, ecology, ethology, and bio- are sterile, or that they are incapable of hybridizing at diversity; in an era in which threats to the natural all. Such an impression ultimately derives from the def- world and its biodiversity are accelerating, it affects inition by Mayr (1940), whereby species are “groups of conservation strategies (Rojas, 1992). -

The Behavioral Ecology of the Tibetan Macaque
Fascinating Life Sciences Jin-Hua Li · Lixing Sun Peter M. Kappeler Editors The Behavioral Ecology of the Tibetan Macaque Fascinating Life Sciences This interdisciplinary series brings together the most essential and captivating topics in the life sciences. They range from the plant sciences to zoology, from the microbiome to macrobiome, and from basic biology to biotechnology. The series not only highlights fascinating research; it also discusses major challenges associ- ated with the life sciences and related disciplines and outlines future research directions. Individual volumes provide in-depth information, are richly illustrated with photographs, illustrations, and maps, and feature suggestions for further reading or glossaries where appropriate. Interested researchers in all areas of the life sciences, as well as biology enthu- siasts, will find the series’ interdisciplinary focus and highly readable volumes especially appealing. More information about this series at http://www.springer.com/series/15408 Jin-Hua Li • Lixing Sun • Peter M. Kappeler Editors The Behavioral Ecology of the Tibetan Macaque Editors Jin-Hua Li Lixing Sun School of Resources Department of Biological Sciences, Primate and Environmental Engineering Behavior and Ecology Program Anhui University Central Washington University Hefei, Anhui, China Ellensburg, WA, USA International Collaborative Research Center for Huangshan Biodiversity and Tibetan Macaque Behavioral Ecology Anhui, China School of Life Sciences Hefei Normal University Hefei, Anhui, China Peter M. Kappeler Behavioral Ecology and Sociobiology Unit, German Primate Center Leibniz Institute for Primate Research Göttingen, Germany Department of Anthropology/Sociobiology University of Göttingen Göttingen, Germany ISSN 2509-6745 ISSN 2509-6753 (electronic) Fascinating Life Sciences ISBN 978-3-030-27919-6 ISBN 978-3-030-27920-2 (eBook) https://doi.org/10.1007/978-3-030-27920-2 This book is an open access publication. -

Controlled Animals
Environment and Sustainable Resource Development Fish and Wildlife Policy Division Controlled Animals Wildlife Regulation, Schedule 5, Part 1-4: Controlled Animals Subject to the Wildlife Act, a person must not be in possession of a wildlife or controlled animal unless authorized by a permit to do so, the animal was lawfully acquired, was lawfully exported from a jurisdiction outside of Alberta and was lawfully imported into Alberta. NOTES: 1 Animals listed in this Schedule, as a general rule, are described in the left hand column by reference to common or descriptive names and in the right hand column by reference to scientific names. But, in the event of any conflict as to the kind of animals that are listed, a scientific name in the right hand column prevails over the corresponding common or descriptive name in the left hand column. 2 Also included in this Schedule is any animal that is the hybrid offspring resulting from the crossing, whether before or after the commencement of this Schedule, of 2 animals at least one of which is or was an animal of a kind that is a controlled animal by virtue of this Schedule. 3 This Schedule excludes all wildlife animals, and therefore if a wildlife animal would, but for this Note, be included in this Schedule, it is hereby excluded from being a controlled animal. Part 1 Mammals (Class Mammalia) 1. AMERICAN OPOSSUMS (Family Didelphidae) Virginia Opossum Didelphis virginiana 2. SHREWS (Family Soricidae) Long-tailed Shrews Genus Sorex Arboreal Brown-toothed Shrew Episoriculus macrurus North American Least Shrew Cryptotis parva Old World Water Shrews Genus Neomys Ussuri White-toothed Shrew Crocidura lasiura Greater White-toothed Shrew Crocidura russula Siberian Shrew Crocidura sibirica Piebald Shrew Diplomesodon pulchellum 3. -
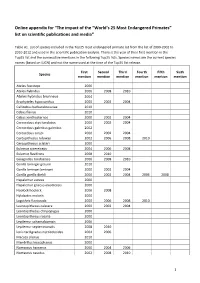
Online Appendix for “The Impact of the “World's 25 Most Endangered
Online appendix for “The impact of the “World’s 25 Most Endangered Primates” list on scientific publications and media” Table A1. List of species included in the Top25 most endangered primate list from the list of 2000-2002 to 2010-2012 and used in the scientific publication analysis. There is the year of their first mention in the Top25 list and the consecutive mentions in the following Top25 lists. Species names are the current species names (based on IUCN) and not the name used at the time of the Top25 list release. First Second Third Fourth Fifth Sixth Species mention mention mention mention mention mention Ateles fusciceps 2006 Ateles hybridus 2006 2008 2010 Ateles hybridus brunneus 2004 Brachyteles hypoxanthus 2000 2002 2004 Callicebus barbarabrownae 2010 Cebus flavius 2010 Cebus xanthosternos 2000 2002 2004 Cercocebus atys lunulatus 2000 2002 2004 Cercocebus galeritus galeritus 2002 Cercocebus sanjei 2000 2002 2004 Cercopithecus roloway 2002 2006 2008 2010 Cercopithecus sclateri 2000 Eulemur cinereiceps 2004 2006 2008 Eulemur flavifrons 2008 2010 Galagoides rondoensis 2006 2008 2010 Gorilla beringei graueri 2010 Gorilla beringei beringei 2000 2002 2004 Gorilla gorilla diehli 2000 2002 2004 2006 2008 Hapalemur aureus 2000 Hapalemur griseus alaotrensis 2000 Hoolock hoolock 2006 2008 Hylobates moloch 2000 Lagothrix flavicauda 2000 2006 2008 2010 Leontopithecus caissara 2000 2002 2004 Leontopithecus chrysopygus 2000 Leontopithecus rosalia 2000 Lepilemur sahamalazensis 2006 Lepilemur septentrionalis 2008 2010 Loris tardigradus nycticeboides -

Analysis of 100 High-Coverage Genomes from a Pedigreed Captive Baboon Colony
Downloaded from genome.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Resource Analysis of 100 high-coverage genomes from a pedigreed captive baboon colony Jacqueline A. Robinson,1 Saurabh Belsare,1 Shifra Birnbaum,2 Deborah E. Newman,2 Jeannie Chan,2 Jeremy P. Glenn,2 Betsy Ferguson,3,4 Laura A. Cox,5,6 and Jeffrey D. Wall1 1Institute for Human Genetics, University of California, San Francisco, California 94143, USA; 2Department of Genetics, Texas Biomedical Research Institute, San Antonio, Texas 78245, USA; 3Division of Genetics, Oregon National Primate Research Center, Beaverton, Oregon 97006, USA; 4Department of Molecular and Medical Genetics, Oregon Health and Science University, Portland, Oregon 97239, USA; 5Center for Precision Medicine, Department of Internal Medicine, Section of Molecular Medicine, Wake Forest School of Medicine, Winston-Salem, North Carolina 27101, USA; 6Southwest National Primate Research Center, Texas Biomedical Research Institute, San Antonio, Texas 78245, USA Baboons (genus Papio) are broadly studied in the wild and in captivity. They are widely used as a nonhuman primate model for biomedical studies, and the Southwest National Primate Research Center (SNPRC) at Texas Biomedical Research Institute has maintained a large captive baboon colony for more than 50 yr. Unlike other model organisms, however, the genomic resources for baboons are severely lacking. This has hindered the progress of studies using baboons as a model for basic biology or human disease. Here, we describe a data set of 100 high-coverage whole-genome sequences obtained from the mixed colony of olive (P. anubis) and yellow (P. cynocephalus) baboons housed at the SNPRC. -

High-Ranking Geladas Protect and Comfort Others After Conflicts
www.nature.com/scientificreports OPEN High-Ranking Geladas Protect and Comfort Others After Conficts Elisabetta Palagi1, Alessia Leone1, Elisa Demuru1 & Pier Francesco Ferrari2 Post-confict afliation is a mechanism favored by natural selection to manage conficts in animal Received: 2 January 2018 groups thus avoiding group disruption. Triadic afliation towards the victim can reduce the likelihood Accepted: 30 August 2018 of redirection (benefts to third-parties) and protect and provide comfort to the victim by reducing its Published: xx xx xxxx post-confict anxiety (benefts to victims). Here, we test specifc hypotheses on the potential functions of triadic afliation in Theropithecus gelada, a primate species living in complex multi-level societies. Our results show that higher-ranking geladas provided more spontaneous triadic afliation than lower- ranking subjects and that these contacts signifcantly reduced the likelihood of further aggression on the victim. Spontaneous triadic afliation signifcantly reduced the victim’s anxiety (measured by scratching), although it was not biased towards kin or friends. In conclusion, triadic afliation in geladas seems to be a strategy available to high-ranking subjects to reduce the social tension generated by a confict. Although this interpretation is the most parsimonious one, it cannot be totally excluded that third parties could also be afected by the negative emotional state of the victim thus increasing a third party’s motivation to provide comfort. Therefore, the debate on the linkage between third-party afliation and emotional contagion in monkeys remains to be resolved. Conficts in social animals can have various immediate and long-term outcomes. Immediately following a con- fict, opponents may show a wide range of responses, from tolerance and avoidance of open confict, to aggres- sion1. -

Patterns of Cranial Shape Variation in the Papionini (Primates
Michelle Singleton Patterns of cranial shape variation in the Department of Anatomy, Papionini (Primates: Cercopithecinae) Midwestern University, 555 31st Street, Downers Grove, Traditional classifications of the Old World monkey tribe Papionini Illinois 60515, U.S.A. (Primates: Cercopithecinae) recognized the mangabey genera E-mail: Cercocebus and Lophocebus as sister taxa. However, molecular studies [email protected] have consistently found the mangabeys to be diphyletic, with Cercocebus and Mandrillus forming a clade to the exclusion of all other Received 10 January 2001 papionins. Recent studies have identified cranial and postcranial Revision received features which distinguish the Cercocebus–Mandrillus clade, however 26 October 2001 and the detailed similarities in cranial shape between the mangabey accepted 12 December 2001 genera are more difficult to reconcile with the molecular evidence. ff Keywords: Papionini, Given the large size di erential between members of the papionin molecular clades, it has frequently been suggested that allometric mangabey diphyly, cranial ff homoplasy, allometry, e ects account for homoplasy in papionin cranial form. A combina- geometric morphometrics. tion of geometric morphometric, bivariate, and multivariate methods was used to evaluate the hypothesis that allometric scaling contributes to craniofacial similarities between like-sized papionin taxa. Patterns of allometric and size-independent cranial shape variation were subsequently described and related to known papionin phylogenetic relationships and patterns of development. Results confirm that allometric scaling of craniofacial shape characterized by positive facial allometry and negative neurocranial allometry is present across adult papionins. Pairwise comparisons of regression lines among genera revealed considerable homogeneity of scaling within the Papionini, however statistically significant differ- ences in regression lines also were noted. -

A Comparison of the Karyotype of Five Species in Genus Macaca
© 2006 The Japan Mendel Society Cytologia 71(2): 161–167, 2006 A Comparison of the Karyotype of Five Species in Genus Macaca (Primate, Cercopithecidae) in Thailand by Using Conventional Staining, G-banding and High-Resolution Technique Alongkoad Tanomtong*,1, Sumpars Khunsook1, Wiwat Kaensa1 and Ruengwit Bunjongrat2 1 Genetics Program, Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen, 40002, Thailand 2 Genetics Program, Department of Botany, Faculty of Science, Chulalongkorn University, Phayathai, Bangkok, 10330, Thailand Received February 7, 2006; accepted March 1, 2006 Summary Cytogenetics of 5 macaque species from genus Macaca in Thailand were studied using lymphocyte cultures and high-resolution techniques. Their chromosome numbers are 2nϭ42, 20 pairs of autosome, and 1 pair of sex-chromosome. M. arctoides and M. mulatta have a fundamental number (NF) 84 in male and female but the others have 83 in male and 84 in female. They have 6 large, 4 medium, 8 small metacentric chromosomes and 8 large, 12 medium, 2 small submetacentric chromosomes respectively. M. fascicularis and M. mulatta have medium metacentric X chromo- some. M. assamensis, M. nemestrina and M. arctoides have medium submetacentric X chromosome. M. arctoides has small submetacentric Y chromosomes, M. mulatta has small metacentric Y chro- mosomes, while M. assamensis, M. fascicularis and M. nemestrina have small telocentric Y chromo- somes. By using G-banding in metaphase and high-resolution technique in late prophase, the results show that the bands are 274, 273, 273, 275, 273 and 351, 350, 350, 352, 350 respectively. Their auto- some and X chromosome are similar but their Y chromosome is different. -

The Sex Lives of Female Olive Baboons (Papio Anubis)
Competition, coercion, and choice: The sex lives of female olive baboons (Papio anubis) DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Jessica Terese Walz Graduate Program in Anthropology The Ohio State University 2016 Dissertation Committee: Dawn M. Kitchen, Chair Douglas E. Crews W. Scott McGraw Copyrighted by Jessica Walz 2016 Abstract Since Darwin first described his theory of sexual selection, evolutionary biologists have used this framework to understand the potential for morphological, physiological, and behavioral traits to evolve within each sex. Recently, researchers have revealed important nuances in effects of sexual coercion, intersexual conflict, and sex role reversals. Among our closest relatives living in complex societies in which individuals interact outside of just the context of mating, the sexual and social lives of individuals are tightly intertwined. An important challenge to biological anthropologists is demonstrating whether female opportunities for mate choice are overridden by male- male competitive and male-female coercive strategies that dominate multi-male, multi- female societies. In this dissertation, I explore interactions between these various mechanisms of competition, coercion, and choice acting on the lives of female olive baboons to determine how they may influence expression of female behavioral and vocal signals, copulatory success with specific males, and the role of female competition in influencing mating patterns. I found females solicit specific males around the time of ovulation. Although what makes some males more preferred is less clear, there is evidence females choose males who might be better future protectors – males who will have long group tenures and are currently ascending the hierarchy.