UNIVERSITY of CALIFORNIA, IRVINE the Resource Acquisition
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Status of the Pacific Spiny Dogfish Shark Resource Off the Continental U.S
Agenda Item G.5 Attachment 3 (Electronic Only) June 2021 DRAFT Disclaimer: These materials do not constitute a formal publication and are for information only. They are in a pre-review, pre-decisional state and should not be formally cited or reproduced. They are to be considered provisional and do not represent any determination or policy of NOAA or the Department of Commerce. Status of the Pacific Spiny Dogfish shark resource off the continental U.S. Pacific Coast in 2021 by Vladlena Gertseva1, Ian Taylor1, John Wallace1, and Sean E. Matson2 1Northwest Fisheries Science Center National Marine Fisheries Service National Oceanic and Atmospheric Administration Seattle, Washington 98112, USA 2West Coast Region National Marine Fisheries Service National Oceanic and Atmospheric Administration Seattle, Washington 98115, USA 2021 1 This report may be cited as: Gertseva, V. Taylor, I.G., Wallace, J.R., Matson, S.E. 2021. Status of the Pacific Spiny Dogfish shark resource off the continental U.S. Pacific Coast in 2021. Pacific Fishery Management Council, Portland, OR. Available from http://www.pcouncil.org/groundfish/stock-assessments/ 2 Table of Contents: Acronyms used in this document .............................................................................................. 6 Executive Summary................................................................................................................. 7 Stock .................................................................................................................................. -

The Ventricles
Guest Editorial Evolution of the Ventricles Solomon Victor, FRCS, FRCP We studied the evolution of ventricles by macroscopic examination of the hearts of Vijaya M. Nayak, MS marine cartilaginous and bony fish, and by angiocardiography and gross examination of Raveen Rajasingh, MPhil the hearts of air-breathing freshwater fish, frogs, turtles, snakes, and crocodiles. A right-sided, thin-walled ventricular lumen is seen in the fish, frog, turtle, and snake. In fish, there is external symmetry of the ventricle, internal asymmetry, and a thick- walled left ventricle with a small inlet chamber. In animals such as frogs, turtles, and snakes, the left ventricle exists as a small-cavitied contractile sponge. The high pressure generated by this spongy left ventricle, the direction of the jet, the ventriculoarterial ori- entation, and the bulbar spiral valve in the frog help to separate the systemic and pul- monary circulations. In the crocodile, the right aorta is connected to the left ventricle, and there is a complete interventricular septum and an improved left ventricular lumen when compared with turtles and snakes. The heart is housed in a rigid pericardial cavity in the shark, possibly to protect it from changing underwater pressure. The pericardial cavity in various species permits move- ments of the heart-which vary depending on the ventriculoarterial orientation and need for the ventricle to generate torque or spin on the ejected blood- that favor run-off into the appropriate arteries and their branches. In the lower species, it is not clear whether the spongy myocardium contributes to myocardial oxygenation. In human beings, spongy myocardium constitutes a rare form of congenital heart disease. -

ESS 345 Ichthyology
ESS 345 Ichthyology Evolutionary history of fishes 12 Feb 2019 (Who’s birthday?) Quote of the Day: We must, however, acknowledge, as it seems to me, that man with all his noble qualities... still bears in his bodily frame the indelible stamp of his lowly origin._______, (1809-1882) Evolution/radiation of fishes over time Era Cenozoic Fig 13.1 Fishes are the most primitive vertebrate and last common ancestor to all vertebrates They start the branch from all other living things with vertebrae and a cranium Chordata Notochord Dorsal hollow nerve cord Pharyngeal gill slits Postanal tail Urochordata Cephalochordata Craniates (mostly Vertebrata) Phylum Chordata sister is… Echinodermata Synapomorphy – They are deuterostomes Fish Evolutionary Tree – evolutionary innovations in vertebrate history Sarcopterygii Chondrichthyes Actinopterygii (fish) For extant fishes Osteichthyes Gnathostomata Handout Vertebrata Craniata Figure only from Berkeley.edu Hypothesis of fish (vert) origins Background 570 MYA – first large radiation of multicellular life – Fossils of the Burgess Shale – Called the Cambrian explosion Garstang Hypothesis 1928 Neoteny of sessile invertebrates Mistake that was “good” Mudpuppy First Vertebrates Vertebrates appear shortly after Cambrian explosion, 530 MYA – Conodonts Notochord replaced by segmented or partially segmented vertebrate and brain is enclosed in cranium Phylogenetic tree Echinoderms, et al. Other “inverts” Vertebrate phyla X Protostomes Deuterostomes Nephrozoa – bilateral animals First fishes were jawless appearing -

Reproductive Biology of the Bonnethead (Sphyrna Tiburo) from the Southeastern U.S
University of North Florida UNF Digital Commons UNF Graduate Theses and Dissertations Student Scholarship 2014 Reproductive Biology of the Bonnethead (Sphyrna tiburo) from the Southeastern U.S. Atlantic Coast Melissa I. Gonzalez De Acevedo University of North Florida, [email protected] Follow this and additional works at: https://digitalcommons.unf.edu/etd Part of the Biology Commons, and the Ecology and Evolutionary Biology Commons Suggested Citation Gonzalez De Acevedo, Melissa I., "Reproductive Biology of the Bonnethead (Sphyrna tiburo) from the Southeastern U.S. Atlantic Coast" (2014). UNF Graduate Theses and Dissertations. 534. https://digitalcommons.unf.edu/etd/534 This Master's Thesis is brought to you for free and open access by the Student Scholarship at UNF Digital Commons. It has been accepted for inclusion in UNF Graduate Theses and Dissertations by an authorized administrator of UNF Digital Commons. For more information, please contact Digital Projects. © 2014 All Rights Reserved REPRODUCTIVE BIOLOGY OF THE BONNETHEAD (SPHYRNA TIBURO) FROM THE SOUTHEASTERN U.S. ATLANTIC COAST by Melissa Gonzalez De Acevedo A thesis submitted to the Department of Biology in partial fulfillment of the requirements for the degree of Masters of Science in Biology UNIVERSITY OF NORTH FLORIDA COLLEGE OF ARTS AND SCIENCES December 2014 Unpublished work, © Melissa Gonzalez De Acevedo CERTIFICATE OF APPROVAL The thesis “Reproductive biology of the bonnethead (Sphyrna tiburo) from the southeastern U.S. Atlantic coast” submitted by Melissa Gonzalez De Acevedo Approved by the thesis committee: Date Dr. Jim Gelsleichter Committee Chair Dr. Carolyn Belcher Dr. Eric Johnson Accepted for the Department of Biology: Dr. Cliff Ross Assistant Chair Accepted for the College of Arts and Sciences: Dr. -

Florida's Fintastic Sharks and Rays Lesson and Activity Packet
Florida's Fintastic Sharks and Rays An at-home lesson for grades 3-5 Produced by: This educational workbook was produced through the support of the Indian River Lagoon National Estuary Program. 1 What are sharks and rays? Believe it or not, they’re a type of fish! When you think “fish,” you probably picture a trout or tuna, but fishes come in all shapes and sizes. All fishes share the following key characteristics that classify them into this group: Fishes have the simplest of vertebrate hearts with only two chambers- one atrium and one ventricle. The spine in a fish runs down the middle of its back just like ours, making fish vertebrates. All fishes have skeletons, but not all fish skeletons are made out of bones. Some fishes have skeletons made out of cartilage, just like your nose and ears. Fishes are cold-blooded. Cold-blooded animals use their environment to warm up or cool down. Fins help fish swim. Fins come in pairs, like pectoral and pelvic fins or are singular, like caudal or anal fins. Later in this packet, we will look at the different types of fins that fishes have and some of the unique ways they are used. 2 Placoid Ctenoid Ganoid Cycloid Hard protective scales cover the skin of many fish species. Scales can act as “fingerprints” to help identify some fish species. There are several different scale types found in bony fishes, including cycloid (round), ganoid (rectangular or diamond), and ctenoid (scalloped). Cartilaginous fishes have dermal denticles (Placoid) that resemble tiny teeth on their skin. -

Seafood Watch Seafood Report
Seafood Watch Seafood Report Sharks and Dogfish With a focus on: Blacktip shark (Carcharhinus limbatus) Common thresher shark (Alopias vulpinus) Dusky smoothhound/smooth dogfish (Mustelus canis) Sandbar shark (Carcharhinus plumbeus) Shortfin mako shark (Isurus oxyrinchus) Spiny dogfish (Squalus acanthias) © Monterey Bay Aquarium Final Report December 21, 2005 Stock Status Update June 9, 2011 Santi Roberts Fisheries Research Analyst Monterey Bay Aquarium SeafoodWatch® Sharks & DogfishReport June 9, 2010 About Seafood Watch® and the Seafood Reports Monterey Bay Aquarium’s Seafood Watch® program evaluates the ecological sustainability of wild-caught and farmed seafood commonly found in the United States marketplace. Seafood Watch® defines sustainable seafood as originating from sources, whether wild-caught or farmed, which can maintain or increase production in the long-term without jeopardizing the structure or function of affected ecosystems. Seafood Watch® makes its science-based recommendations available to the public in the form of regional pocket guides that can be downloaded from the Internet (seafoodwatch.org) or obtained from the Seafood Watch® program by emailing [email protected]. The program’s goals are to raise awareness of important ocean conservation issues and empower seafood consumers and businesses to make choices for healthy oceans. Each sustainability recommendation on the regional pocket guides is supported by a Seafood Report. Each report synthesizes and analyzes the most current ecological, fisheries and ecosystem science on a species, then evaluates this information against the program’s conservation ethic to arrive at a recommendation of “Best Choices,” “Good Alternatives,” or “Avoid.” The detailed evaluation methodology is available upon request. In producing the Seafood Reports, Seafood Watch® seeks out research published in academic, peer-reviewed journals whenever possible. -

Ground Sharks
click for previous page - v - TABLE OF CONTENTS Code Page 9. ORDER CARCHARHINIFORMES - GROUND SHARKS ....................................................................................... 251 9.1 FAMILY SCYLIORHINIDAE - Catsharks .................................................. SCYL ........................................... 253 Apristurus....................................................................................................... SCYL Aprist ................................ 257 A. atlanticus ..................................................................................... SCYL Aprist 1 ............................... 261 A. brunneus ...................................................................................... SCYL Aprist 2 ............................... 262 A. canutus ............................................................................................ SCYL Aprist 3 ............................... 263 A. herklotsi ........................................................................................ SCYL Aprist 4 ............................... 264 A. indicus ............................................................................................. SCYL Aprist 5 ............................... 265 A. investigatoris ................................................................................... SCYL Aprist 6 ............................... 267 A. japonicus ....................................................................................... SCYL Aprist 7 ............................... 268 -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -

Last Meal Reflects Spiral-Shaped Intestine 6 January 2016
Last meal reflects spiral-shaped intestine 6 January 2016 performed statistical analyses that revealed unknown patterns of distribution of the intestinal structure", explains Marcelo Sánchez. All of this led to the finding that Saurichthys had a straight stomach and a spiral valve. Energetic lifestyle The findings by the Zurich-based paleontologists unveil previously unexpected convergences with the gastrointestinal anatomy of present-day sharks and rays. "The anatomy of the gastrointestinal tract of Saurichthys, in particular the many windings in Extinct predatory fish Saurichthys. Credit: UZH the spiral valve, show the most original digestion organs of earlier fishes", says Thodoris Argyriou, a PhD student at the Paleontological Institute and Museum of the University of Zurich. The spiral A last meal provides new insights: The fossilized valve of Saurichthys indicates an energy-laden food remains of the extinct predatory fish lifestyle that was adjusted to the predatory and Saurichthys reflect its spiral-shaped intestine. The reproductive behavior of the genus. "The large spiral valve in fossils from Southern Switzerland is number of windings increased the surface area for similar to that of sharks and rays. Paleontologists digestion, which is sure to have provided the fish from the University of Zurich have thus closed a with more energy", says Thodoris Argyriou. gap in the knowledge concerning the evolution of the gastrointestinal tract in vertebrates. More information: Thodoris Argyriou et al. Exceptional preservation reveals gastrointestinal A last supper has provided some interesting anatomy and evolution in early actinopterygian findings. The digested and undigested remains of fishes, Scientific Reports (2016). DOI: the last meal eaten by a Saurichthys, a Triassic 10.1038/srep18758 bony fish, were discovered in a extraordinary case of preservation that paleontologists at the University of Zurich were quick to take advantage of. -

Fishes Scales & Tails Scale Types 1
Phylum Chordata SUBPHYLUM VERTEBRATA Metameric chordates Linear series of cartilaginous or boney support (vertebrae) surrounding or replacing the notochord Expanded anterior portion of nervous system THE FISHES SCALES & TAILS SCALE TYPES 1. COSMOID (most primitive) First found on ostracaderm agnathans, thick & boney - composed of: Ganoine (enamel outer layer) Cosmine (thick under layer) Spongy bone Lamellar bone Perhaps selected for protection against eurypterids, but decreased flexibility 2. GANOID (primitive, still found on some living fish like gar) 3. PLACOID (old scale type found on the chondrichthyes) Dentine, tooth-like 4. CYCLOID (more recent scale type, found in modern osteichthyes) 5. CTENOID (most modern scale type, found in modern osteichthyes) TAILS HETEROCERCAL (primitive, still found on chondrichthyes) ABBREVIATED HETEROCERCAL (found on some primitive living fish like gar) DIPHYCERCAL (primitive, found on sarcopterygii) HOMOCERCAL (most modern, found on most modern osteichthyes) Agnatha (class) [connect the taxa] Cyclostomata (order) Placodermi Acanthodii (class) (class) Chondrichthyes (class) Osteichthyes (class) Actinopterygii (subclass) Sarcopterygii (subclass) Dipnoi (order) Crossopterygii (order) Ripidistia (suborder) Coelacanthiformes (suborder) Chondrostei (infra class) Holostei (infra class) Teleostei (infra class) CLASS AGNATHA ("without jaws") Most primitive - first fossils in Ordovician Bottom feeders, dorsal/ventral flattened Cosmoid scales (Ostracoderms) Pair of eyes + pineal eye - present in a few living fish and reptiles - regulates circadian rhythms Nine - seven gill pouches No paired appendages, medial nosril ORDER CYCLOSTOMATA (60 spp) Last living representatives - lampreys & hagfish Notochord not replaced by vertebrae Cartilaginous cranium, scaleless body Sea lamprey predaceous - horny teeth in buccal cavity & on tongue - secretes anti-coaggulant Lateral Line System No stomach or spleen 5 - 7 year life span - adults move into freshwater streams, spawn, & die. -

Thermoregulation Strategies of Deep Diving Ectothermic Sharks
THERMOREGULATION STRATEGIES OF DEEP DIVING ECTOTHERMIC SHARKS A DISSERTATION SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAIʻI AT MĀNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCOTOR OF PHILOSOPHY IN ZOOLOGY (MARINE BIOLOGY) AUGUST 2020 By. Mark A. Royer Dissertation Committee: Kim Holland, Chairperson Brian Bowen Carl Meyer Andre Seale Masato Yoshizawa Keywords: Ectothermic, Thermoregulation, Biologging, Hexanchus griseus, Syphrna lewini, Shark ACKNOWLEDGEMENTS Thank you to my advisor Dr. Kim Holland and to Dr. Carl Meyer for providing me the privilege to pursue a doctoral degree in your lab, which provided more experiences and opportunities than I could have ever imagined. The research environment you provided allowed me to pursue new frontiers in the field and take on challenging questions. Thank you to my committee members Dr. Brian Bowen, Dr. Andre Seale, and Dr. Masato Yoshizawa, for providing your ideas, thoughts, suggestions, support and encouragement through the development of my dissertation. I would like to give my sincere thanks to all of my committee members and to the Department of Biology for taking their time to provide their support and accommodation as I finished my degree during a rather unprecedented and uncertain time. I am very grateful to everyone at the HIMB Shark Lab including Dr. Melanie Hutchinson, Dr. James Anderson, Jeff Muir, and Dr. Daniel Coffey. I learned so much from all of you and we have shared several lifetimes worth of experiences. Thank you to Dr. James Anderson for exciting side projects we have attempted and will continue to pursue in the future. Thank you to Dr. -
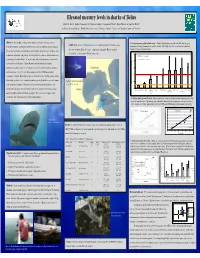
Mercury in Sharks of Belize, 2008
Elevated mercury levels in sharks of Belize David C. Evers 1, Rachel T. Graham 2, Neil Hammerschlag 3, Christopher Perkins 4, Robert Michener 5, and Tim Divoll 1 BioDiversity Research Institute 1, Wildlife Conservation Society 2, University of Miami 3, University of Connecticut 4, and Boston University 5. Abstract: Mercury (Hg) loading in global aquatic ecosystems is a growing concern. 1. Mercury exposure profile by shark species - Highest Hg levels were recorded in the bull, blacktip, great Study Area : A total of 14 distinct locations were sampled in the Gulf of Honduras along Compelling evidence of widespread adverse effects in fish and wildlife populations indicates hammerhead, scalloped hammerhead, and nurse sharks. Lowest Hg levels were recorded in the bonnethead, the coast of southern Belize (red circle). Comparison shark muscle Hg concentrations sharpnose, lemon and sandbar sharks. the rate of transformation to methylmercury is problematic. Long-lived, apex predators such from the U.S. are from southern Florida (white circle). 2.5 as sharks are at high risk to Hg toxicity. We investigated the occurrence of Hg in sharks from USEPA Advisory = 0.3 ug/g, ww coastal waters of southern Belize. In our pilot study, 101 sharks representing 9 species were FDA & WHO Advisory = 1.0 ug/g, ww 2.0 analyzed for muscle Hg levels. Highest Hg levels were recorded in bull, blacktip, hammerhead, and nurse sharks. Lowest Hg levels were recorded in bonnethead, sharpnose, 1.5 and lemon sharks. Over 88% of the sharks sampled exceeded USEPA human health 1.0 consumption standards. Muscle Hg strongly correlated with size for blacktip and nurse sharks.